Hi,
i have two problem when i process a dataset of a small membrane protein. First, The 3D refinement all(homogenous heterogenous or non uniform refinement) give a FSC like below. All parameters are default.
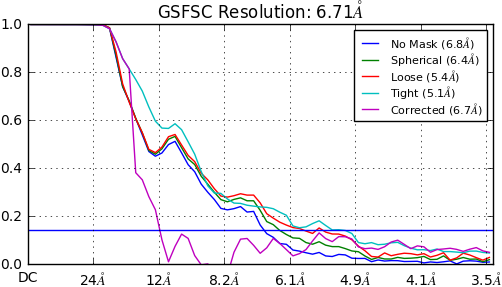
The reconstruction doesn’t get better after 3D refinement. I wonder if this curve is due to the particles haven’t optimize to its right orientation parameters?
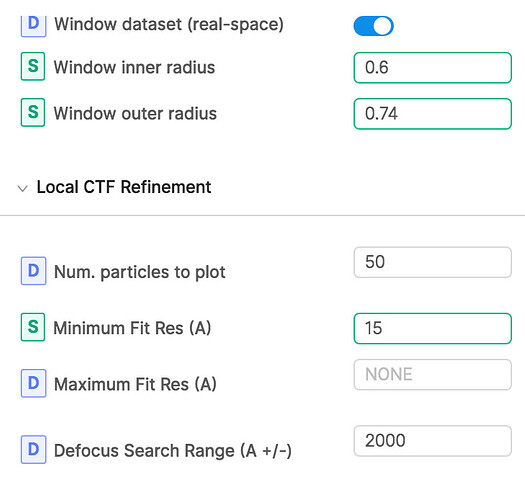
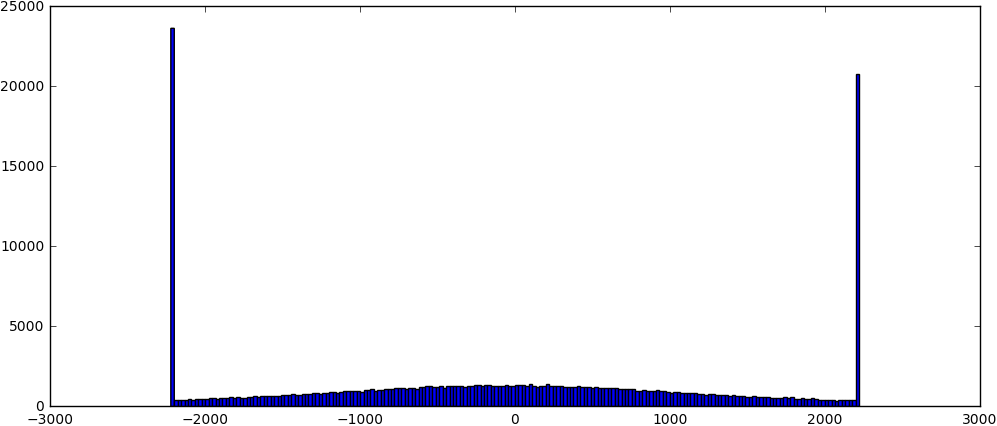
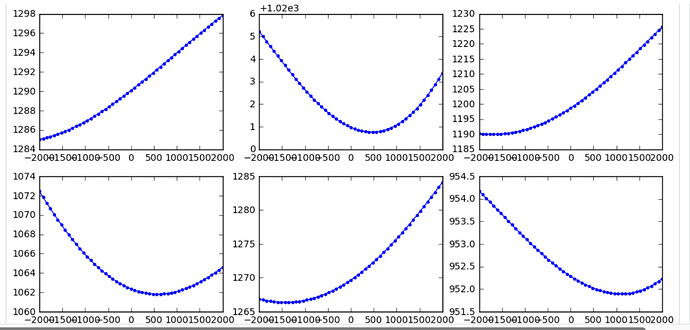
Second, i try to do per particle CTF refinement. But the result seems not very confident or accurate like the description in Tutorial: CTF Refinement. Below is the parameter and result. I don’t know if this is because i didn’t use right parameter or it is due to the data itself.
Thanks in advance.
Lei
Hi Lei,
You probably don’t want to play with ctf refinement until you have a good starting refinement, probably at 4.5A or better. I see similar fsc curves for my small asymmetric membrane protein where the micelle is dominating the signal and there are few other features to aid alignment. In both cases, I suspect the ab initio model is not correct and the particle set needs some pruning with 3D classification in relion. I have had trouble using cryosparc’s heterogeneous refinement for data like this.
One place to start might be adjusting the initial and maximum resolutions during ab initio and using more classes. With this strategy you can try to separate different classes from junk, say by putting the max res to 6A or 8A.
Also, why did you adjust the window inner/outer radius? How big is the particle in relation to the box size?
-Aaron
Thanks very much for your suggestion. Yes, the particle set may needs further pruning. Because i use this window inner/outer radius during 2D classfication which is better than the default parameter. Thus, I keep this window inner/outer radius in all following processes. But i think it doesn’t play much role. The original box size is about twice the length of particle diameter.
Best,
Lei