Hi, I have a dataset of movies with 60 frames. And I did Patch Motion with all frames (J1) and 40 frames (J2). When I got particles from J1 micrographs, can I re-extract the same particles from J2? That means extracting particles from J1 micrographs and J2 micrographs with the same particle locations. I tried to extract directly from J2 micrographs and particles locations from J1 micrographs, but it didn’t work. Any suggests will be appreciated. Thank you!
Jianhao
1 Like
August 2021 Update: The instructions below are no longer required for cryoSPARC v3.2 or newer. Instead, create a new extraction job, connect the particles from the first picking job and connect the exposures from the second motion correction job.
Hi @jianhaoc, yes you can do this but it’s a bit complicated. The particle locations are tied to the path of the motion-corrected micrographs on disk. That means you’ll have to duplicate the particle locations and manually modify the “path” component of the location. Here’s how to do that.
-
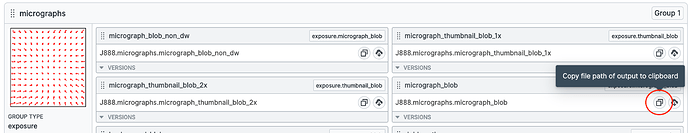
Copy the micrograph_blob path from the J2 outputs as indicated. Note this down somewhere
-
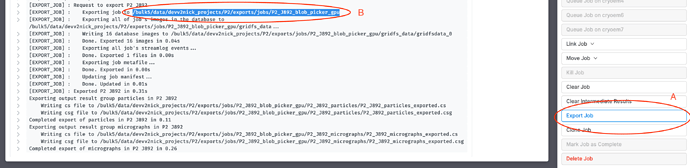
Export the job you used to generate the particle locations from the micrographs in J1 (A). Note down the path of the exported job (B), as indicated (I’m using a Blob Picker job for this example)
The directory structure of this exported path should look something like this (assuming J3 is the job that generated the particles)
/path/to/project/exports/jobs/P1_J3_blob_picker_gpu
├── events.bson
├── gridfs_data
│ └── gridfsdata_0
├── job.json
├── P1_J3_micrographs
│ ├── J1
│ ├── J2
│ ├── P1_J3_micrographs_exported.cs
│ └── P1_J3_micrographs_exported.csg
└── P1_J3_particles
├── J1
├── P1_J3_particles_exported.cs
└── P1_J3_particles_exported.csg
Note the full path to the exported particles data file (e.g., P1_J3_particles_exported.cs)
-
Paste the following code into a text editor and make the substitutions below
from cryosparc_compute.dataset import Dataset
micrographs_path = '<J2 MICROGRAPHS PATH HERE>'
exported_particles_path = '<J1 PARTICLES CS PATH HERE>'
micrographs = Dataset().from_file(micrographs_path)
particles = Dataset().from_file(exported_particles_path)
mic_uid_to_path = dict(zip(micrographs.data['uid'], micrographs.data['micrograph_blob/path']))
for particle in particles.get_items():
particle['location/micrograph_path'] = mic_uid_to_path[particle['location/micrograph_uid']]
particles.to_file(exported_particles_path)
- Change
<J2 MICROGRAPHS PATH HERE> to the J2 micrograph blob path from Step 1
- Change
<J1 PARTICLES CS PATH HERE> to the full path of the exported particles data file from Step 2 (e.g., /path/to/project/exports/jobs/P1_J3_blob_picker_gpu/P1_J3_particles/P1_J3_particles_exported.cs)
- If you’re running cryoSPARC v2.16 or below, change
cryosparc_compute to cryosparc2_compute
-
Open a terminal with access to the cryosparcm command line interface. Enter cryosparcm icli to enter interactive CLI mode
-
Paste the substituted code from Step 3 into the prompt and press Enter to run it. Check for any errors in the output before proceeding
-
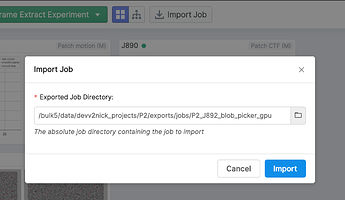
Use the “Import Job” button to re-import the exported job with the now-modified particle locations
You may see an error notification, which you may safely ignore
-
Extract the particles from J1 as you normally would
-
Extract the particles from J2: Create an Extract Particles job, connect the micrographs directly from J2 and the particle locations from the imported job at Step 6. Run the job
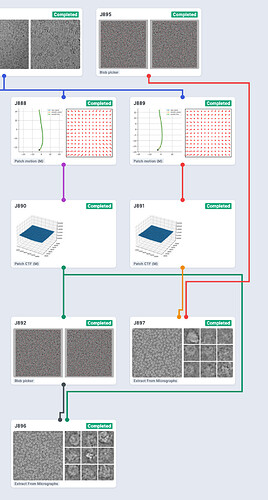
Here’s the resulting workspace. J895 is the modified, re-imported blob-picker job. Note that I also ran Patch CTF on the micrographs before picking the particles and connected those micrographs to the Extract jobs
Hope that helps, let me know if you run into any trouble at some point.
1 Like
Thank you for the details.
Just trying to follow this @nfrasser - is there any prospect for making this less complicated in a future version? This is a lot of mucking about to just test a different per frame dose, different gain, frame range etc, when one is working with the same set of movies…
It would also be great if there was the capacity to access the alignment parameters after patch motion, so one could test different doses, gain references, or calculate per frame reconstructions without having to completely redo motion correction
Cheers
Oli
@olibclarke in the latest cryoSPARC v3.2 you should now be able to extract the same particles by connecting the Blob Picks from the first motion correction result and the exposures from the second motion correction results.
I’ll bring up enabling access to alignment parameters with the rest of team.
1 Like
Thanks @nfrasser - does this work with all pickers, refined particle sets etc, or just blob picker? Because when I try to do this with particles from a refinement job, it doesn’t work - it thinks there are 0 particles on every micrograph (at least with the CPU extraction job).
Or maybe this was introduced in a patch? I am on v3.2.0+210629, next to latest patch I believe
@olibclarke it should work with most pickers and particle sets so long as the micrographs from where the particles were originally picked all came from the same Import job. If the micrographs were, say, re-imported after outside processing then they’ll have different unique IDs and be considered different from the original ones.
I also know we have some edge-cases with Topaz and Deep Picker picks where this may still be an issue.
Are any of these the case for your setup?
Patch v3.2.0+210629 should be sufficient, but let me double check.
Hi @nfrasser,
These were re-imported, in order to use a different gain reference during Patch Motion, but they are the same set of movies. Yes, I originally picked using Topaz.
Would it be possible to have some facility to ignore unique IDs in such a case, or copy them across from an existing dataset which has movies with identical names?
Oli
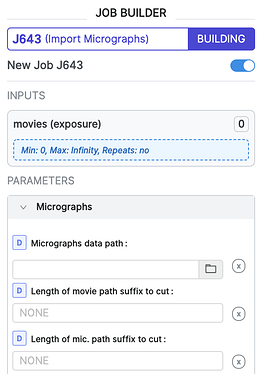
The only way to do this right now is if you still have the original movies imported in cryoSPARC: You can match the newly-imported micrographs by specifying them in the movies input under the Import Micrographs job.
The micrograph file names must closely match the movie names for this to work - use the Length of movie path suffix to cut and Length of mic. path suffix to cut parameters to augment the filenames and ensure a match.
to be clear, I am not importing micrographs, I am importing movies, to apply a new gain in patch motion. Or are you suggesting I then “re-import” the patch motion corrected mics?
@nfrasser for this to work with V3.2, do the motion correction jobs have to be perfomed in the v3.2 ? I tried to run an extraction from the motion correction job (performed in V2) but it completes with zero particle output.