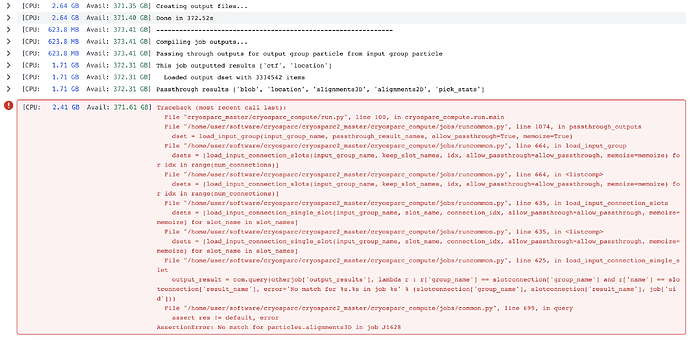
Here you go:
Traceback (most recent call last):
File "cryosparc_master/cryosparc_compute/run.py", line 100, in cryosparc_compute.run.main
File "/home/user/software/cryosparc/cryosparc2_master/cryosparc_compute/jobs/runcommon.py", line 1074, in passthrough_outputs
dset = load_input_group(input_group_name, passthrough_result_names, allow_passthrough=True, memoize=True)
File "/home/user/software/cryosparc/cryosparc2_master/cryosparc_compute/jobs/runcommon.py", line 664, in load_input_group
dsets = [load_input_connection_slots(input_group_name, keep_slot_names, idx, allow_passthrough=allow_passthrough, memoize=memoize) for idx in range(num_connections)]
File "/home/user/software/cryosparc/cryosparc2_master/cryosparc_compute/jobs/runcommon.py", line 664, in <listcomp>
dsets = [load_input_connection_slots(input_group_name, keep_slot_names, idx, allow_passthrough=allow_passthrough, memoize=memoize) for idx in range(num_connections)]
File "/home/user/software/cryosparc/cryosparc2_master/cryosparc_compute/jobs/runcommon.py", line 635, in load_input_connection_slots
dsets = [load_input_connection_single_slot(input_group_name, slot_name, connection_idx, allow_passthrough=allow_passthrough, memoize=memoize) for slot_name in slot_names]
File "/home/user/software/cryosparc/cryosparc2_master/cryosparc_compute/jobs/runcommon.py", line 635, in <listcomp>
dsets = [load_input_connection_single_slot(input_group_name, slot_name, connection_idx, allow_passthrough=allow_passthrough, memoize=memoize) for slot_name in slot_names]
File "/home/user/software/cryosparc/cryosparc2_master/cryosparc_compute/jobs/runcommon.py", line 625, in load_input_connection_single_slot
output_result = com.query(otherjob['output_results'], lambda r : r['group_name'] == slotconnection['group_name'] and r['name'] == slotconnection['result_name'], error='No match for %s.%s in job %s' % (slotconnection['group_name'], slotconnection['result_name'], job['uid']))
File "/home/user/software/cryosparc/cryosparc2_master/cryosparc_compute/jobs/common.py", line 699, in query
assert res != default, error
AssertionError: No match for particles.alignments3D in job J1628
The job was cloned from a previous job and the inputs swapped out, but the previous job was created in the same version of cryoSPARC a few days before.