We love using cryoSPARC Live, and have some instances where we would like to use the Patch aligned micrographs for import into other programs. I can see how to ‘export exposures’ from Live into my usual cryoSPARC instance. From there however, I am unclear on how to export the aligned, average micrographs.
Thanks!
Hi @simonbrown,
When you select “Export Exposures” in the cryoSPARC Live configuration tab, you should see this:
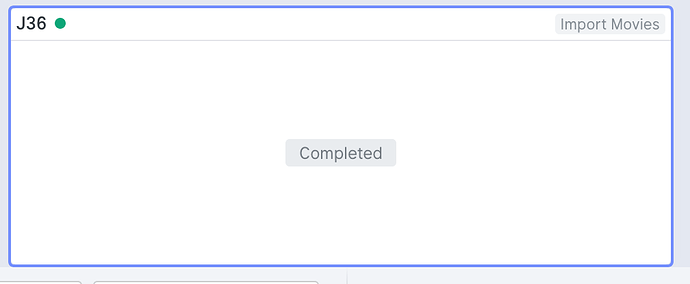
This means that Live created an “Import Movies” job in the cryoSPARC Project Workspace:
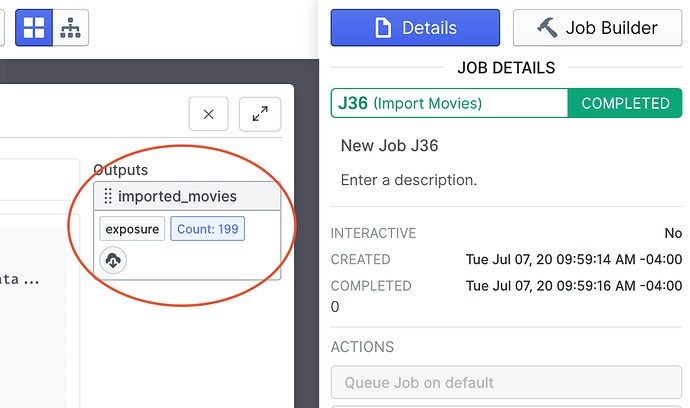
This input group includes references to all data that Live computed for every imported movie, including raw movie frames, motion-corrected micrograph and CTF.
You can use this output by dragging it into the input slot of any other job that supports it as you normally would in regular cryoSPARC.
G’day Nick,
Thanks for the reply. I think my question wasn’t entirely clear. I want to be able to output the aligned movies for use in other packages.
What I do currently is dig into the cryosparc database and copy out the aligned .mrc files from the ‘S1’ motion corrected directory. It would be nice to have a GUI option to access these files for export.
Thanks!
Hi @simonbrown, gotcha, thank you for clarifying. If I understand correctly, but this:
You mean a function to download the files from the browser running the cryoSPARC web interface to your local machine? Or do you need a way to copy the MRC files from the S1 directory to a different location on the machine running cryoSPARC?
We wouldn’t be able to do first case due to how large the MRC files are; it would be much slower than the process you’re already using (I’m assuming something like the scp and rsync tools).
This seems the more useful approach. Now I know where the files are it isn’t as pressing of an issue, but for a new user of cryoSPARC live this would be helpful. I can imagine a function under the configuration tab in Live, where as well as the current ‘export’ buttons, (that export into cryoSPARC), another set of buttons that can create a new export directory in the relevant Pxx directory, and place the aligned micrographs here.