Hi,
I run a ‘Reconstruct only’ job and got half A(J**_volume_map_half_A.mrc), half B(J**_volume_map_half_B.mrc), and a mask(J**_volume_mask_fsc.mrc).
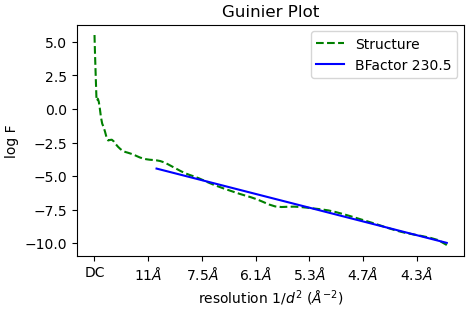
In the ‘Reconstruct only’ job, the estimated Bfactor is -89.8:
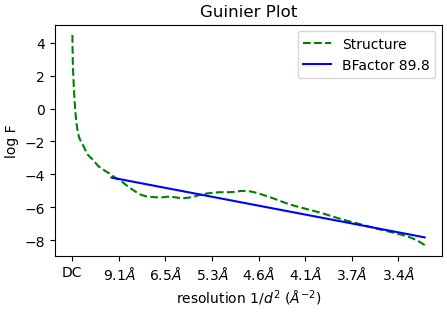
But if I use RELION for post-processing with the same half maps and mask as input, the estimated B factor differs by a factor of two compared to CryoSPARC:
relion_postprocess --i J214_volume_map_half_A.mrc --i2 J214_volume_map_half_B.mrc --o relion_sharp --angpix 0.644 --mask J214_volume_mask_fsc.mrc --auto_bfac true
It reports:
== Reading input half-reconstructions:
+ half1-map: J214_volume_map_half_A.mrc
+ half2-map: J214_volume_map_half_B.mrc
+ --mtf_angpix was not provided, assuming pixel size in raw micrographs is the same as in particles, 0.644 A/px.
== Using a user-provided mask ...
+ input mask: J214_volume_mask_fsc.mrc
+ fraction f (solvent mask based): 35.6179
+ molecular weight inside protein mask: 259965
== Masking input maps ...
+ randomize phases beyond: 14.4256 Angstroms
== Applying sqrt(2*FSC/(FSC+1)) weighting (as in Rosenthal & Henderson, 2003) ...
== Fitting straight line through Guinier plot to find B-factor ...
+ fit from resolution: 10
+ fit until resolution: 0
+ slope of fit: -11.6472
+ intercept of fit: -10.8523
+ correlation of fit: 0.792205
+ apply b-factor of: -46.5887
== Writing output files ...
+ Processed map: relion_sharp.mrc
+ Processed masked map: relion_sharp_masked.mrc
+ Metadata file: relion_sharp.star
+ FINAL RESOLUTION: 3.24171
With the same half maps and mask, RELION apply b-factor of -46.5887, while CryoSPARC apply a b-factor of -89.8.
I wonder why does estimated Bfactor in CryoSPARC differ by a factor of two compared to RELION. (My cryoSPARC is v4.1.0)
BTW, in the source code of RELION, I can see that RELION divide high frequency part by e^{-0.25 b x^2}:
void applyBFactorToMap(MultidimArray<Complex > &FT, int ori_size, RFLOAT bfactor, RFLOAT angpix)
{
FOR_ALL_ELEMENTS_IN_FFTW_TRANSFORM(FT)
{
int r2 = kp * kp + ip * ip + jp * jp;
RFLOAT res = sqrt((RFLOAT)r2)/(ori_size * angpix); // get resolution in 1/Angstrom
if (res <= 1. / (angpix * 2.) ) // Apply B-factor sharpening until Nyquist, then low-pass filter later on (with a soft edge)
{
DIRECT_A3D_ELEM(FT, k, i, j) *= exp( -(bfactor / 4.) * res * res);
}
else
{
DIRECT_A3D_ELEM(FT, k, i, j) = 0.;
}
}
}
I wonder how whether the coefficient for B-factor sharpening in CryoSPARC is the same as that used by RELION.
Any advice would help.
Thank you very much!