Dear Everyone,
I am experincing some problems analyzing a dataset. My sample is a dimeric membrane complex of 105Å depth (45 Å in membrane), 205 Å length, and 110 Å width.
I manage to obtain good initia 2D classes, even though I can’t see any internal features. From there after multiple rounds of Ab-initio and 2D clasees I get a 3D volume that looks like my complex of interest, but then I get really confusing results in the homogeneous and NU-refinements that I have done.
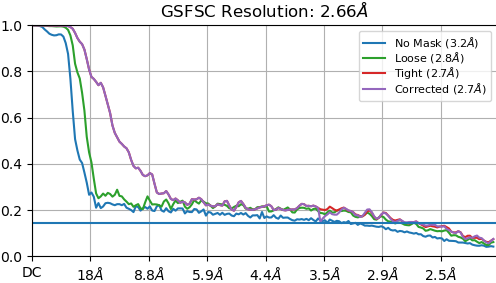
First of all I don’t fully understand the shape of this FSC, what might the issue be? Moreover, when I look at the map, the surface looks really sharp, but the inside is “empty” and I can see no features at all. (I would have sent more pictures but the forum is not allowing me)
At this point I am very confused if the problem is sample related or processing realted. Would a bigger dataset help in this this?
One of the possible explanation I’m thinking of is that in my sample might be present a contamination 10% of another membrane complex, a circular trimer with a diameter also of 200 Å length. And this is confusing everything from the 2D classification. Is this a silly idea? if not, do you have any suggestion on how to try to detangle the particles that belong to different complexes? I have already tried heterogeneous refinement, ab inition with a large number of classes and some other things.
Sorry for the long post and thanks to anyone that can help,
Giovanni
Hi Giovanni,
How did you pick your particles?
Looking at this FSC, the first thing I would suggest as a possibility is that you may have some duplicate particles. Have you tried using Remove Duplicates?
Cheers
Oli
1 Like
Hi Olib,
Thanks for your help, you really answered solved my problem! Remove Duplicates got rid of around 1k particles and now I have a nomal looking FSC. I’m still really a novice and without much guidance, so sorry for the big mistake.
On the other side of things, with a 10Å reconsctruction now I’m still missing density inside my reconsctruction, is it just because the resolution is too low or this problem is originating somewhere else?
all the best,
Giovanni
1 Like
Hi Giovanni,
Hard to say without more details. Can you see secondary structural details in your 2D classes? If not you may want to optimize picking (or go back and optimize the sample further). Does the ab initio model look sensible, does it have recognizable structural features? Can you fit an initial model to your map?
Cheers
Oli
Hi Oli,
I will try to optimize picking, since I can see just dome helices in some of the 2D classes, I feel like optimizing the sample from the biochemical point of view is the way to go, the side view parallel to the membrane look very similar for my two complexes, both in size and shape.
My ab initio model tough looks sensible, I can distinguish the main features and I can fit my comples pretty well.
thanks for all your help, you’ve been great,
All the best,
Giovanni
