Hi,
thanks for the quick answer!
I tried it adding --movies flag, but am getting an “MicrographOriginalPixelSize” error.
Traceback (most recent call last):
File "/home/perun-d12/anaconda3/envs/pyem2/lib/python3.11/site-packages/pandas/core/indexes/base.py", line 3653, in get_loc
return self._engine.get_loc(casted_key)
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
File "pandas/_libs/index.pyx", line 147, in pandas._libs.index.IndexEngine.get_loc
File "pandas/_libs/index.pyx", line 176, in pandas._libs.index.IndexEngine.get_loc
File "pandas/_libs/hashtable_class_helper.pxi", line 7080, in pandas._libs.hashtable.PyObjectHashTable.get_item
File "pandas/_libs/hashtable_class_helper.pxi", line 7088, in pandas._libs.hashtable.PyObjectHashTable.get_item
KeyError: 'rlnMicrographOriginalPixelSize'
The above exception was the direct cause of the following exception:
Traceback (most recent call last):
File "/media/longstorage/Tadej/cryocloud/datasets/enc_raw/cs/csparc2star.py", line 179, in <module>
sys.exit(main(parser.parse_args()))
^^^^^^^^^^^^^^^^^^^^^^^^^
File "/media/longstorage/Tadej/cryocloud/datasets/enc_raw/cs/csparc2star.py", line 59, in main
data_general = metadata.cryosparc_2_cs_movie_parameters(cs, passthroughs=pts, trajdir=trajdir, path=args.micrograph_path)
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
File "/media/longstorage/Tadej/cryocloud/cs2star/pyem/pyem/metadata/cryosparc2.py", line 224, in cryosparc_2_cs_movie_parameters
data_general[star.Relion.MICROGRAPHPIXELSIZE] / data_general[star.Relion.MICROGRAPHORIGINALPIXELSIZE]
~~~~~~~~~~~~^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
File "/home/perun-d12/anaconda3/envs/pyem2/lib/python3.11/site-packages/pandas/core/frame.py", line 3761, in __getitem__
indexer = self.columns.get_loc(key)
^^^^^^^^^^^^^^^^^^^^^^^^^
File "/home/perun-d12/anaconda3/envs/pyem2/lib/python3.11/site-packages/pandas/core/indexes/base.py", line 3655, in get_loc
raise KeyError(key) from err
KeyError: 'rlnMicrographOriginalPixelSize'
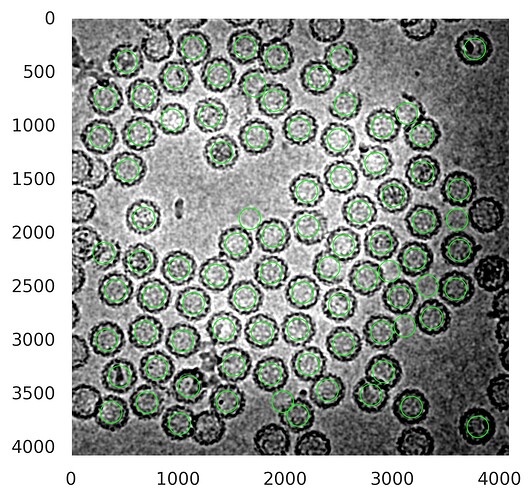
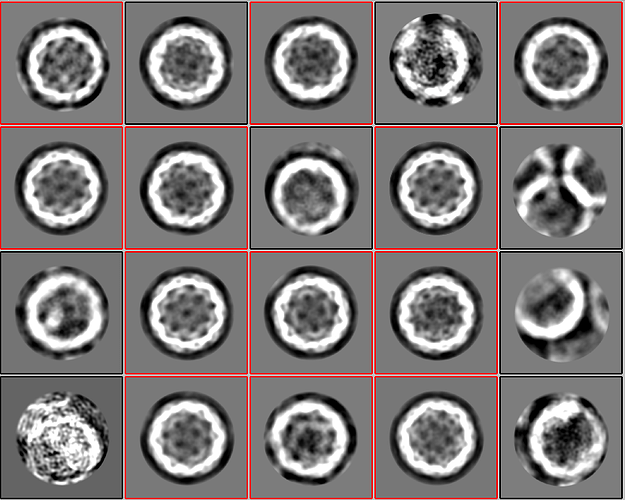
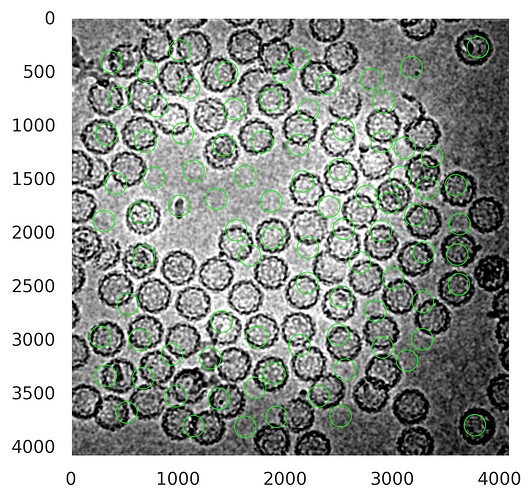
For the second question. I’m doing both 2D classifications in RELION. One is with particles extracted in CryoSparc, and for the second one, I re-extract the same particles in Relion. In theory, extracted particles should be the same, however, I am getting very different results.
2D classification of particles from Cryosparc:
2D classification of particles from Relion (same particle locations!):
Not sure what seems to be the problem. Thanks for all the help!