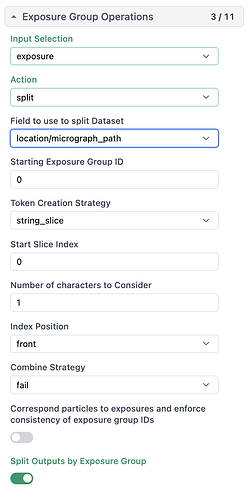
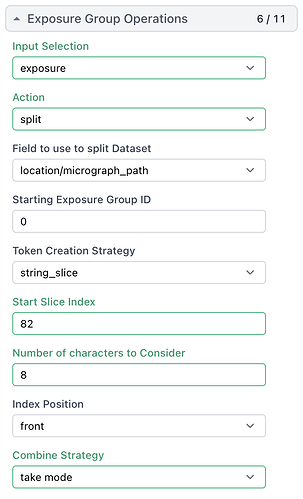
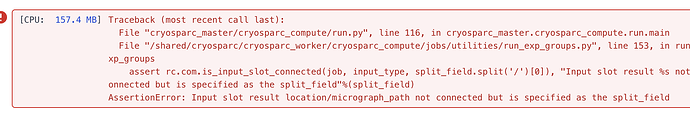
Hi All,
Thanks for your responses! There is currently no easy way to do this via the GUI, unless you import each set of movies by gridsquare (setting the exposure group ID parameter for each set in the import job), reprocess the mics through Patch CTF estimation, and then reassign particles to those micrographs. This is definitely less than ideal, therefore @nfrasser made a cs-tools script that would be able to accomplish what you seek to do @wjnicol
To use this script (located at the bottom of my message):
- Ensure you have a working and version matched cryosparc-tools environment.
- Copy script from below and save as
split_exposures_by_grid_square.py and ensure it has the correct permissions to run.
- Change the license, email, password, host, and base_port values in the script to match your instance.
- Launch script from the command line using the following command
python split_exposures_by_grid_square.py P1 W2 J3 accepted_exposures where:
- Project is the project containing the Live Exposure Export
- Workspace is the workspace you would like the split exposure groups to be placed
- Job is the Live Export Exposures job from your CS-live session
- ‘accepted_exposures’ is the output group from the Live Export Exposures job (this does not change)
If you wanted to use the script on the outputs of a Patch CTF Estimation job, then you would need to use the correct project and job for the Patch CTF Estimation job and then change the output group to exposures.
Additionally, we have noted a feature request for maintaining full path info in some format such that analyses like these can be performed within the GUI.
Please let me know if you have any issues.
Best,
Kye
# e.g,. python split_exposures_by_grid_square.py P3 W4 J42 accepted_exposures
import sys
from pathlib import Path
from cryosparc.tools import CryoSPARC
# Parse arguments
assert len(sys.argv) == 5, f"Usage: python {sys.argv[0]} <PROJECT-ID> <WORKSPACE-ID> <MOVIES-JOB-ID> <MOVIES-OUTPUT-NAME>"
project_uid, workspace_uid, job_uid, input_name = sys.argv[1:]
# Connect to CryoSPARC
cs = CryoSPARC( # SUBSTITUTE CRYOSPARC INSTANCE DETAILS HERE
license="<LICENSE ID>",
email="<EMAIL>",
password="<PASSWORD>",
host="<HOST NAME>",
base_port=<PORT NUMBER>,
)
assert cs.test_connection()
# Load entities
project = cs.find_project(project_uid)
workspace = project.find_workspace(workspace_uid)
job = project.find_job(job_uid)
movies = job.load_output(input_name, slots=["movie_blob"])
# Split up movies dataset by resolving symlinks
print(f"Splitting {len(movies)} exposures by grid square folder...")
project_dir = Path(project.dir())
grid_square_idxs: dict[str, list[int]] = {}
for i, link_path in enumerate(movies["movie_blob/path"]):
link_path_abs = project_dir / str(link_path)
movie_path_abs = link_path_abs.resolve()
if movie_path_abs.parent.name != "Data":
print(f"WARNING: Movie is not in a GridSquare/Data folder: {movie_path_abs}", file=sys.stderr)
continue
# Resolve original movie path and add to a grid square index group
grid_square_dir_path = movie_path_abs.parent.parent
if grid_square_dir_path.name not in grid_square_idxs:
print(f"Found grid square folder {grid_square_dir_path.name}")
grid_square_idxs[grid_square_dir_path.name] = []
grid_square_idxs[grid_square_dir_path.name].append(i)
assert grid_square_idxs, f"ERROR: Selected exposures output has no movies or no matching movies with the correct absolute path format"
# Create external job and add a group for each grid square
print(f"Saving {len(grid_square_idxs)} outputs job...")
for grid_square, idxs in grid_square_idxs.items():
saved_job_uid = workspace.save_external_result(
movies.take(idxs),
type="exposure",
name=grid_square,
slots=["movie_blob"],
passthrough=(job_uid, input_name),
title=f"Exposures for {grid_square}",
)
print(f"Saved {grid_square} to {saved_job_uid}")