Here we go:
[csparc@biomix ~]$ cryosparcm eventlog P5 J515 | head -n 40
[Sun, 14 Jul 2024 14:57:30 GMT] License is valid.
[Sun, 14 Jul 2024 14:57:30 GMT] Launching job on lane biomix target biomix ...
[Sun, 14 Jul 2024 14:57:30 GMT] Launching job on cluster biomix
[Sun, 14 Jul 2024 14:57:30 GMT]
====================== Cluster submission script: ========================
==========================================================================
#!/bin/bash
#SBATCH --job-name=cryosparc_P5_J515
#SBATCH --partition=cryosparc
#SBATCH --output=/mnt/parashar/cspark_files/CS-sm74/J515/job.log
#SBATCH --error=/mnt/parashar/cspark_files/CS-sm74/J515/job.log
#SBATCH --nodes=1
#SBATCH --mem=16000M
#SBATCH --ntasks-per-node=1
#SBATCH --cpus-per-task=6
#SBATCH --gres=gpu:1
#SBATCH --gres-flags=enforce-binding
srun /usr/localMAIN/cryosparc/cryosparc_worker/bin/cryosparcw run --project P5 --job J515 --master_hostname biomix.dbi.udel.edu --master_command_core_port 39002 > /mnt/parashar/cspark_files/CS-sm74/J515/job.log 2>&1
==========================================================================
==========================================================================
[Sun, 14 Jul 2024 14:57:30 GMT] -------- Submission command:
sbatch /mnt/parashar/cspark_files/CS-sm74/J515/queue_sub_script.sh
[Sun, 14 Jul 2024 14:57:30 GMT] -------- Cluster Job ID:
679604
[Sun, 14 Jul 2024 14:57:30 GMT] -------- Queued on cluster at 2024-07-14 10:57:30.657604
[Sun, 14 Jul 2024 14:57:31 GMT] -------- Cluster job status at 2024-07-14 10:57:31.069857 (0 retries)
JOBID PARTITION NAME USER ST TIME NODES NODELIST(REASON)
679604 cryosparc cryospar csparc R 0:01 1 biomix10
[Sun, 14 Jul 2024 14:57:32 GMT] [CPU RAM used: 92 MB] Job J515 Started
[Sun, 14 Jul 2024 14:57:32 GMT] [CPU RAM used: 92 MB] Master running v4.5.3, worker running v4.5.3
[Sun, 14 Jul 2024 14:57:32 GMT] [CPU RAM used: 92 MB] Working in directory: /mnt/parashar/cspark_files/CS-sm74/J515
[Sun, 14 Jul 2024 14:57:32 GMT] [CPU RAM used: 92 MB] Running on lane biomix
[Sun, 14 Jul 2024 14:57:32 GMT] [CPU RAM used: 92 MB] Resources allocated:
[Sun, 14 Jul 2024 14:57:32 GMT] [CPU RAM used: 92 MB] Worker: biomix
[Sun, 14 Jul 2024 14:57:32 GMT] [CPU RAM used: 92 MB] CPU : [0, 1, 2, 3, 4, 5]
[Sun, 14 Jul 2024 14:57:32 GMT] [CPU RAM used: 92 MB] GPU : [0]
[Sun, 14 Jul 2024 14:57:32 GMT] [CPU RAM used: 92 MB] RAM : [0, 1]
[Sun, 14 Jul 2024 14:57:32 GMT] [CPU RAM used: 92 MB] SSD : False
[Sun, 14 Jul 2024 14:57:32 GMT] [CPU RAM used: 92 MB] --------------------------------------------------------------
[Sun, 14 Jul 2024 14:57:32 GMT] [CPU RAM used: 92 MB] Importing job module for job type patch_motion_correction_multi...
Traceback (most recent call last):
File "<string>", line 9, in <module>
BrokenPipeError: [Errno 32] Broken pipe
command 2
[csparc@biomix ~]$ cryosparcm joblog P5 J515 | tail -n 40
========= sending heartbeat at 2024-07-14 22:02:27.167072
========= sending heartbeat at 2024-07-14 22:02:37.182244
========= sending heartbeat at 2024-07-14 22:02:47.198172
========= sending heartbeat at 2024-07-14 22:02:57.215540
========= sending heartbeat at 2024-07-14 22:03:07.230151
========= sending heartbeat at 2024-07-14 22:03:17.239818
========= sending heartbeat at 2024-07-14 22:03:27.253724
========= sending heartbeat at 2024-07-14 22:03:37.271657
========= sending heartbeat at 2024-07-14 22:03:47.289765
========= sending heartbeat at 2024-07-14 22:03:57.307739
========= sending heartbeat at 2024-07-14 22:04:07.326268
========= sending heartbeat at 2024-07-14 22:04:17.345515
========= sending heartbeat at 2024-07-14 22:04:27.362153
========= sending heartbeat at 2024-07-14 22:04:37.380736
========= sending heartbeat at 2024-07-14 22:04:47.399128
========= sending heartbeat at 2024-07-14 22:04:57.416897
========= sending heartbeat at 2024-07-14 22:05:07.434426
========= sending heartbeat at 2024-07-14 22:05:17.450146
========= sending heartbeat at 2024-07-14 22:05:27.467578
========= sending heartbeat at 2024-07-14 22:05:37.484845
========= sending heartbeat at 2024-07-14 22:05:47.502330
========= sending heartbeat at 2024-07-14 22:05:57.520148
========= sending heartbeat at 2024-07-14 22:06:07.537891
========= sending heartbeat at 2024-07-14 22:06:17.556618
========= sending heartbeat at 2024-07-14 22:06:27.574242
========= sending heartbeat at 2024-07-14 22:06:37.590242
========= sending heartbeat at 2024-07-14 22:06:47.608114
========= sending heartbeat at 2024-07-14 22:06:57.626769
========= sending heartbeat at 2024-07-14 22:07:07.645911
========= sending heartbeat at 2024-07-14 22:07:17.665957
========= heartbeat failed at 2024-07-14 22:07:17.674537:
========= sending heartbeat at 2024-07-14 22:07:27.684655
========= heartbeat failed at 2024-07-14 22:07:27.692649:
========= sending heartbeat at 2024-07-14 22:07:37.702753
========= heartbeat failed at 2024-07-14 22:07:37.710694:
************* Connection to cryosparc command lost. Heartbeat failed 3 consecutive times at 2024-07-14 22:07:37.710743.
/usr/localMAIN/cryosparc/cryosparc_worker/bin/cryosparcw: line 150: 1116721 Killed python -c "import cryosparc_compute.run as run; run.run()" "$@"
slurmstepd-biomix10: error: Detected 1 oom-kill event(s) in StepId=679604.0. Some of your processes may have been killed by the cgroup out-of-memory handler.
srun: error: biomix10: task 0: Out Of Memory
slurmstepd-biomix10: error: Detected 1 oom-kill event(s) in StepId=679604.batch. Some of your processes may have been killed by the cgroup out-of-memory handler.
command 3
[csparc@biomix ~]$ cryosparcm cli "get_job('P5', 'J515', 'version', 'job_type', 'params_spec', 'status', 'instance_information')"
{'_id': '6693e74d03031811dbc0e16c', 'instance_information': {'CUDA_version': '11.8', 'available_memory': '247.14GB', 'cpu_model': 'Intel(R) Xeon(R) Silver 4410Y', 'driver_version': '12.3', 'gpu_info': [{'id': 0, 'mem': 47810936832, 'name': 'NVIDIA L40S', 'pcie': '0000:3d:00'}], 'ofd_hard_limit': 131072, 'ofd_soft_limit': 1024, 'physical_cores': 24, 'platform_architecture': 'x86_64', 'platform_node': 'biomix10', 'platform_release': '5.15.0-105-generic', 'platform_version': '#115-Ubuntu SMP Mon Apr 15 09:52:04 UTC 2024', 'total_memory': '251.55GB', 'used_memory': '1.87GB'}, 'job_type': 'patch_motion_correction_multi', 'params_spec': {'output_fcrop_factor': {'value': '1/2'}}, 'project_uid': 'P5', 'status': 'completed', 'uid': 'J515', 'version': 'v4.5.3'}
command 4
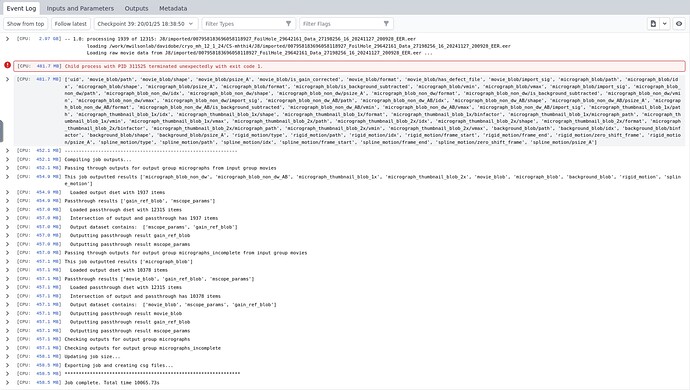
[csparc@biomix ~]$ cryosparcm eventlog P5 J515 | tail -n 40
Writing background estimate to J515/motioncorrected/010265144360898815905_FoilHole_8956123_Data_8936159_8936161_20240709_144445_fractions_background.mrc ...
Done in 0.04s
Writing motion estimates...
Done in 0.01s
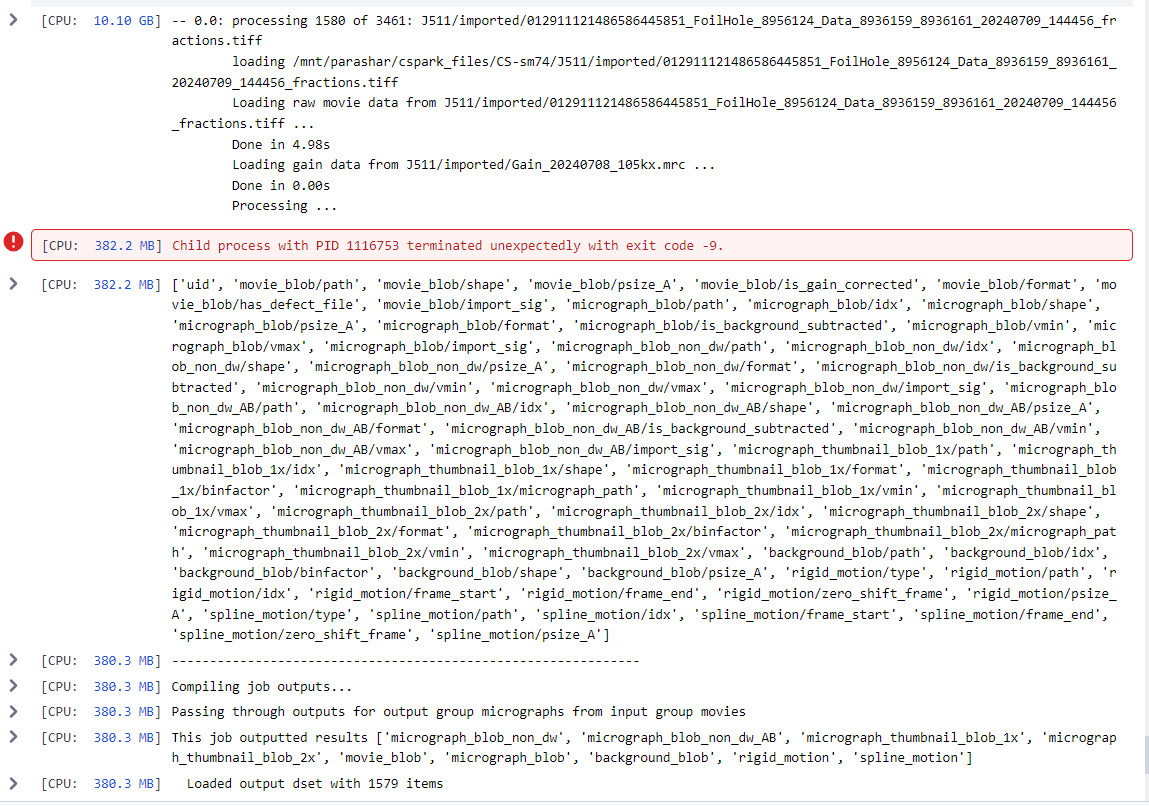
[Sun, 14 Jul 2024 22:06:34 GMT] [CPU RAM used: 10099 MB] -- 0.0: processing 1580 of 3461: J511/imported/012911121486586445851_FoilHole_8956124_Data_8936159_8936161_20240709_144456_fractions.tiff
loading /mnt/parashar/cspark_files/CS-sm74/J511/imported/012911121486586445851_FoilHole_8956124_Data_8936159_8936161_20240709_144456_fractions.tiff
Loading raw movie data from J511/imported/012911121486586445851_FoilHole_8956124_Data_8936159_8936161_20240709_144456_fractions.tiff ...
Done in 4.98s
Loading gain data from J511/imported/Gain_20240708_105kx.mrc ...
Done in 0.00s
Processing ...
[Sun, 14 Jul 2024 22:07:04 GMT] [CPU RAM used: 382 MB] Child process with PID 1116753 terminated unexpectedly with exit code -9.
[Sun, 14 Jul 2024 22:07:04 GMT] [CPU RAM used: 382 MB] ['uid', 'movie_blob/path', 'movie_blob/shape', 'movie_blob/psize_A', 'movie_blob/is_gain_corrected', 'movie_blob/format', 'movie_blob/has_defect_file', 'movie_blob/import_sig', 'micrograph_blob/path', 'micrograph_blob/idx', 'micrograph_blob/shape', 'micrograph_blob/psize_A', 'micrograph_blob/format', 'micrograph_blob/is_background_subtracted', 'micrograph_blob/vmin', 'micrograph_blob/vmax', 'micrograph_blob/import_sig', 'micrograph_blob_non_dw/path', 'micrograph_blob_non_dw/idx', 'micrograph_blob_non_dw/shape', 'micrograph_blob_non_dw/psize_A', 'micrograph_blob_non_dw/format', 'micrograph_blob_non_dw/is_background_subtracted', 'micrograph_blob_non_dw/vmin', 'micrograph_blob_non_dw/vmax', 'micrograph_blob_non_dw/import_sig', 'micrograph_blob_non_dw_AB/path', 'micrograph_blob_non_dw_AB/idx', 'micrograph_blob_non_dw_AB/shape', 'micrograph_blob_non_dw_AB/psize_A', 'micrograph_blob_non_dw_AB/format', 'micrograph_blob_non_dw_AB/is_background_subtracted', 'micrograph_blob_non_dw_AB/vmin', 'micrograph_blob_non_dw_AB/vmax', 'micrograph_blob_non_dw_AB/import_sig', 'micrograph_thumbnail_blob_1x/path', 'micrograph_thumbnail_blob_1x/idx', 'micrograph_thumbnail_blob_1x/shape', 'micrograph_thumbnail_blob_1x/format', 'micrograph_thumbnail_blob_1x/binfactor', 'micrograph_thumbnail_blob_1x/micrograph_path', 'micrograph_thumbnail_blob_1x/vmin', 'micrograph_thumbnail_blob_1x/vmax', 'micrograph_thumbnail_blob_2x/path', 'micrograph_thumbnail_blob_2x/idx', 'micrograph_thumbnail_blob_2x/shape', 'micrograph_thumbnail_blob_2x/format', 'micrograph_thumbnail_blob_2x/binfactor', 'micrograph_thumbnail_blob_2x/micrograph_path', 'micrograph_thumbnail_blob_2x/vmin', 'micrograph_thumbnail_blob_2x/vmax', 'background_blob/path', 'background_blob/idx', 'background_blob/binfactor', 'background_blob/shape', 'background_blob/psize_A', 'rigid_motion/type', 'rigid_motion/path', 'rigid_motion/idx', 'rigid_motion/frame_start', 'rigid_motion/frame_end', 'rigid_motion/zero_shift_frame', 'rigid_motion/psize_A', 'spline_motion/type', 'spline_motion/path', 'spline_motion/idx', 'spline_motion/frame_start', 'spline_motion/frame_end', 'spline_motion/zero_shift_frame', 'spline_motion/psize_A']
[Sun, 14 Jul 2024 22:07:04 GMT] [CPU RAM used: 380 MB] --------------------------------------------------------------
[Sun, 14 Jul 2024 22:07:04 GMT] [CPU RAM used: 380 MB] Compiling job outputs...
[Sun, 14 Jul 2024 22:07:04 GMT] [CPU RAM used: 380 MB] Passing through outputs for output group micrographs from input group movies
[Sun, 14 Jul 2024 22:07:05 GMT] [CPU RAM used: 380 MB] This job outputted results ['micrograph_blob_non_dw', 'micrograph_blob_non_dw_AB', 'micrograph_thumbnail_blob_1x', 'micrograph_thumbnail_blob_2x', 'movie_blob', 'micrograph_blob', 'background_blob', 'rigid_motion', 'spline_motion']
[Sun, 14 Jul 2024 22:07:05 GMT] [CPU RAM used: 380 MB] Loaded output dset with 1579 items
[Sun, 14 Jul 2024 22:07:05 GMT] [CPU RAM used: 380 MB] Passthrough results ['gain_ref_blob', 'mscope_params']
[Sun, 14 Jul 2024 22:07:05 GMT] [CPU RAM used: 381 MB] Loaded passthrough dset with 3461 items
[Sun, 14 Jul 2024 22:07:05 GMT] [CPU RAM used: 381 MB] Intersection of output and passthrough has 1579 items
[Sun, 14 Jul 2024 22:07:05 GMT] [CPU RAM used: 381 MB] Output dataset contains: ['mscope_params', 'gain_ref_blob']
[Sun, 14 Jul 2024 22:07:05 GMT] [CPU RAM used: 381 MB] Outputting passthrough result gain_ref_blob
[Sun, 14 Jul 2024 22:07:05 GMT] [CPU RAM used: 381 MB] Outputting passthrough result mscope_params
[Sun, 14 Jul 2024 22:07:05 GMT] [CPU RAM used: 381 MB] Passing through outputs for output group micrographs_incomplete from input group movies
[Sun, 14 Jul 2024 22:07:05 GMT] [CPU RAM used: 381 MB] This job outputted results ['micrograph_blob']
[Sun, 14 Jul 2024 22:07:05 GMT] [CPU RAM used: 381 MB] Loaded output dset with 1882 items
[Sun, 14 Jul 2024 22:07:05 GMT] [CPU RAM used: 381 MB] Passthrough results ['movie_blob', 'gain_ref_blob', 'mscope_params']
[Sun, 14 Jul 2024 22:07:05 GMT] [CPU RAM used: 381 MB] Loaded passthrough dset with 3461 items
[Sun, 14 Jul 2024 22:07:05 GMT] [CPU RAM used: 381 MB] Intersection of output and passthrough has 1882 items
[Sun, 14 Jul 2024 22:07:05 GMT] [CPU RAM used: 381 MB] Output dataset contains: ['mscope_params', 'gain_ref_blob', 'movie_blob']
[Sun, 14 Jul 2024 22:07:05 GMT] [CPU RAM used: 381 MB] Outputting passthrough result movie_blob
[Sun, 14 Jul 2024 22:07:05 GMT] [CPU RAM used: 381 MB] Outputting passthrough result gain_ref_blob
[Sun, 14 Jul 2024 22:07:05 GMT] [CPU RAM used: 381 MB] Outputting passthrough result mscope_params
[Sun, 14 Jul 2024 22:07:05 GMT] [CPU RAM used: 381 MB] Checking outputs for output group micrographs
[Sun, 14 Jul 2024 22:07:05 GMT] [CPU RAM used: 381 MB] Checking outputs for output group micrographs_incomplete
[Sun, 14 Jul 2024 22:07:05 GMT] [CPU RAM used: 381 MB] Updating job size...
[Sun, 14 Jul 2024 22:07:10 GMT] [CPU RAM used: 382 MB] Exporting job and creating csg files...
[Sun, 14 Jul 2024 22:07:10 GMT] [CPU RAM used: 382 MB] ***************************************************************
[Sun, 14 Jul 2024 22:07:10 GMT] [CPU RAM used: 382 MB] Job complete. Total time 25774.07s