Hi,
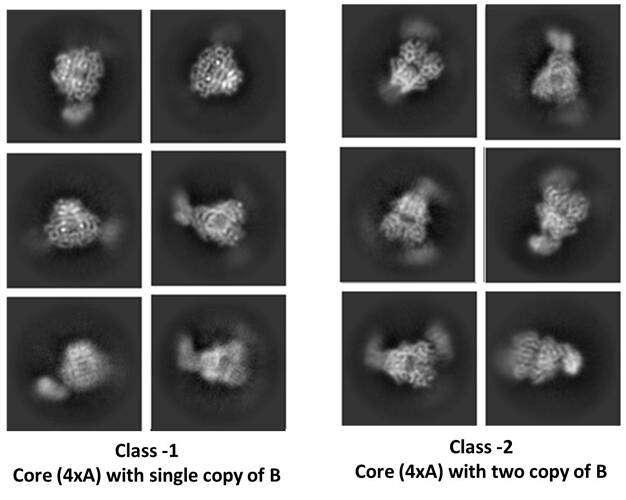
I am new to cryoEM and analysing my first dataset. My proein complex contain two proteins, protein-A forms tetrameric core and protein-B binds to core (either one/two copies only, not sure). Iniital 2D classes shows heterogenous population of mainly two types of particles, 1) tetrameric core of protein-A with single bound copy of protein-B and 2) tetrameric core of protein-A with two bound copies of protein-B (see attached representative 2D class images). My question is for type-2 particles. Is it possible that class-2 particles may be appeared due to simply mis-alignment of class-1 particles, meaning that they are aligned by core but just other way around and appearance of second copy of B in class-2 is simply artifacts due to misalignment. I am asking this question becasue I am not much aware if 2D classification job (program) of cryoSPARC can make such systamatic error? Or such error by programe is rather not possible and presence of two copy of protein-B can be considered as a real-event?
The comment from dev/experienced-users will be appreciated. Thank you!
Hi ravi, this is a common issue when there is a rigid core with floppy or heterogeneous outer density, as you have. I think two things are possibly occurring simultaneously - you could have heterogeneous protein composition, different stoichiometry, as you say. And also protein B is somewhat floppy in relation to the rigid core protein A. Both of these result in alignment based mostly on protein A with some contribution by protein B, but it is a smear. I would extract with a larger box and repeat 2D classification, since protein B is close to the edge.
I am not confident in your counting the copies by eye in 2D. For example the top right class in class 2 looks similar to the middle left class in class 1. Protein B is getting averaged out, either due to conformational variability or compositional heterogeneity.
The 2D classes look quite nice (in my opinion) and you should go to 3D with a larger box to get an idea of what is happening. Try ab inito into multiple classes or heterogeneous refinement into multiple classes. Avoid imposing symmetry for initial refinements as that can enhance the averaging out effect that you see in 2D here. I would work with 1.5-2x larger box and particles binned to 4A for initial 3D work.
Hi user123,
Thank you for providing more insights on this issue. Usage of larger box size is certaily a good idea. I will try it. I agree with you that the second copy is being averaged out might be due to confomrational variabiloty or compositional heterogeneity. But I my specific question now is 'would it be possible to conclude from this classes some particles indeed possess two copy of protein-B, or it can still be artifacts of misalignment of core other way around? Is there any way to check this?
For instance, now I could already get two compositionally different 3D volumes from this dataset, one with single copy and another with two copies of protein-B. How about running heterogenous refinement again using two-copies-particles with both one-copy and two-copies 3D volumes?
Thank you,