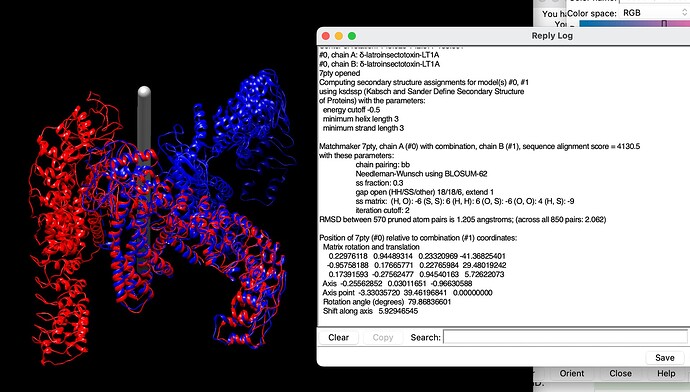
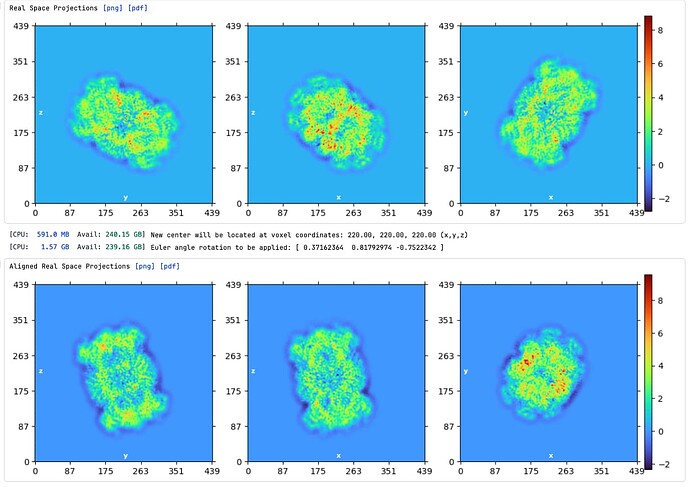
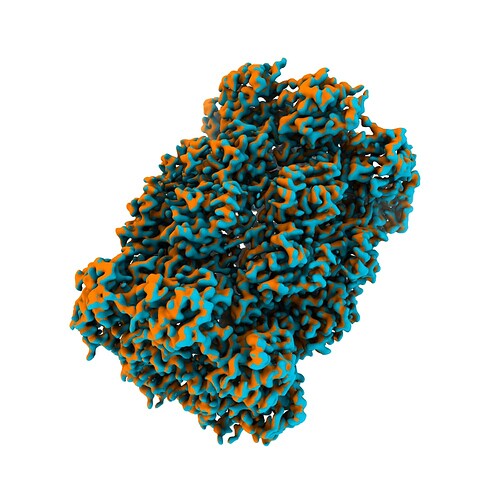
Hi Rich, I tried this, but it gave the following error:
(cryosparc-tools) user@ubuntu:~$ python realign_script.py
Connection succeeded to CryoSPARC command_core at http://localhost:39002
Connection succeeded to CryoSPARC command_vis at http://localhost:39003
Connection succeeded to CryoSPARC command_rtp at http://localhost:39005
Traceback (most recent call last):
File "realign_script.py", line 43, in <module>
rot_mat, translation = parse_chimerax_matrix_string(chimx_mat)
File "realign_script.py", line 30, in parse_chimerax_matrix_string
[
File "realign_script.py", line 31, in <listcomp>
[float(y) for y in x.split(' ')]
File "realign_script.py", line 31, in <listcomp>
[float(y) for y in x.split(' ')]
ValueError: could not convert string to float: ''
Using a slightly edited version of your script:
from cryosparc.tools import CryoSPARC
import json
import numpy as np
from pathlib import Path
from scipy.spatial.transform import Rotation as R
with open(Path('~/cs_instance_info.json').expanduser(), 'r') as f:
instance_info = json.load(f)
cs = CryoSPARC(**instance_info)
assert cs.test_connection()
project_number = "P51"
workspace_number = "W6"
# src should be a symmetry expansion in C1 if you want
# to use the sym_expand/idx later
src_job_number = "J253"
src_job_particles_name = "particles"
src_job_vol_name = "volume"
lane_to_use = "default"
project = cs.find_project(project_number)
workspace = project.find_workspace(workspace_number)
job = project.find_job(src_job_number)
unrotated_particles = job.load_output(src_job_particles_name)
def parse_chimerax_matrix_string(chimx_mat:str) -> np.array:
full_matrix = np.array(
[
[float(y) for y in x.split(' ')]
for x in chimx_mat.split('\n')
]
)
rot_matrix = full_matrix[:,:3]
translation = full_matrix[:, 3].T
return (rot_matrix, translation)
chimx_mat = """ 0.22976117 0.94489315 0.23320967 -41.36825376
-0.95758188 0.17665770 0.22765985 29.48019271
0.17391594 -0.27562475 0.94540163 5.72622007"""
rot_mat, translation = parse_chimerax_matrix_string(chimx_mat)
# we want a new center in voxels, not a translation in A
translation /= unrotated_particles['alignments3D/psize_A'][0]
new_box_center = unrotated_particles['blob/shape'][0][0] / 2 - translation
euler = R.from_matrix(rot_mat).as_euler('ZYZ')
# chimeraX translates *after* rotating, CryoSPARC translates *before*.
# thus, we need two VAT jobs. One rotating, one translating.
unshifted_job = workspace.create_job(
type = "volume_alignment_tools",
connections = {
"particles": (src_job_number, src_job_particles_name),
"volume": (src_job_number, src_job_vol_name)
},
params = {
# yes, this is the right parameter name for the Euler angles
'recenter_axang': ','.join(str(x) for x in euler)
}
)
unshifted_job.queue(lane_to_use)
unshifted_job.wait_for_done()
unshift_id = unshifted_job.doc['uid']
shifted_job = workspace.create_job(
type = "volume_alignment_tools",
connections = {
"particles": (unshift_id, "particles"),
"volume": (unshift_id, "volume")
},
params = {
"recenter_shift": ",".join(str(x) for x in new_box_center)
}
)
shifted_job.queue(lane_to_use)
shifted_job.wait_for_done()
shifted_id = shifted_job.doc['uid']
shifted_results = project.find_job(shifted_id)
shifted_particles = shifted_results.load_output('particles')
shifted_particles['sym_expand/idx'] = 1
project.save_external_result(
workspace_uid=workspace_number,
dataset=shifted_particles,
type='particle',
slots=['sym_expand'],
passthrough=(shifted_id, 'particles'),
title='non-point-group sym expand'
)
Any idea what might be going wrong?
EDIT:
Adding an operation to strip trailing whitespace seemed to fix the issue, although the resulting volume alignment tools job does not have the expected rotation:
from cryosparc.tools import CryoSPARC
import json
import numpy as np
from pathlib import Path
from scipy.spatial.transform import Rotation as R
with open(Path('~/cs_instance_info.json').expanduser(), 'r') as f:
instance_info = json.load(f)
cs = CryoSPARC(**instance_info)
assert cs.test_connection()
project_number = "P51"
workspace_number = "W6"
# src should be a symmetry expansion in C1 if you want
# to use the sym_expand/idx later
src_job_number = "J253"
src_job_particles_name = "particles"
src_job_vol_name = "volume"
lane_to_use = "default"
project = cs.find_project(project_number)
workspace = project.find_workspace(workspace_number)
job = project.find_job(src_job_number)
unrotated_particles = job.load_output(src_job_particles_name)
#def parse_chimerax_matrix_string(chimx_mat:str) -> np.array:
# full_matrix = np.array(
# [
# [float(y) for y in x.split(' ')]
# for x in chimx_mat.split('\n')
# ]
# )
# rot_matrix = full_matrix[:,:3]
# translation = full_matrix[:, 3].T
# return (rot_matrix, translation)
def parse_chimerax_matrix_string(chimx_mat: str) -> np.array:
print("Input string:")
print(chimx_mat)
lines = chimx_mat.strip().split('\n') # Strip leading/trailing whitespace
print("Split lines:")
print(lines)
full_matrix = np.array(
[
[float(y) for y in x.split()] # No need to specify ' ' as split argument, it will split by any whitespace
for x in lines
]
)
print("Parsed matrix:")
print(full_matrix)
rot_matrix = full_matrix[:, :3]
translation = full_matrix[:, 3].T
return rot_matrix, translation
chimx_mat = """ 0.22976117 0.94489315 0.23320967 -41.36825376
-0.95758188 0.17665770 0.22765985 29.48019271
0.17391594 -0.27562475 0.94540163 5.72622007 """
print(chimx_mat)
rot_mat, translation = parse_chimerax_matrix_string(chimx_mat)
# we want a new center in voxels, not a translation in A
translation /= unrotated_particles['alignments3D/psize_A'][0]
new_box_center = unrotated_particles['blob/shape'][0][0] / 2 - translation
euler = R.from_matrix(rot_mat).as_euler('ZYZ')
# chimeraX translates *after* rotating, CryoSPARC translates *before*.
# thus, we need two VAT jobs. One rotating, one translating.
unshifted_job = workspace.create_job(
type = "volume_alignment_tools",
connections = {
"particles": (src_job_number, src_job_particles_name),
"volume": (src_job_number, src_job_vol_name)
},
params = {
# yes, this is the right parameter name for the Euler angles
'recenter_axang': ','.join(str(x) for x in euler)
}
)
unshifted_job.queue(lane_to_use)
unshifted_job.wait_for_done()
unshift_id = unshifted_job.doc['uid']
shifted_job = workspace.create_job(
type = "volume_alignment_tools",
connections = {
"particles": (unshift_id, "particles"),
"volume": (unshift_id, "volume")
},
params = {
"recenter_shift": ",".join(str(x) for x in new_box_center)
}
)
shifted_job.queue(lane_to_use)
shifted_job.wait_for_done()
shifted_id = shifted_job.doc['uid']
shifted_results = project.find_job(shifted_id)
shifted_particles = shifted_results.load_output('particles')
shifted_particles['sym_expand/idx'] = 1
project.save_external_result(
workspace_uid=workspace_number,
dataset=shifted_particles,
type='particle',
slots=['sym_expand'],
passthrough=(shifted_id, 'particles'),
title='non-point-group sym expand'
)
EDIT:
Ok got it to work! The input matrix has to be derived from measure rotation as applied to two duplicate maps - not models fit to the maps. Then, if the origin of both maps has been set to the center of the box, it does the trick! Thanks heaps!