Hi,
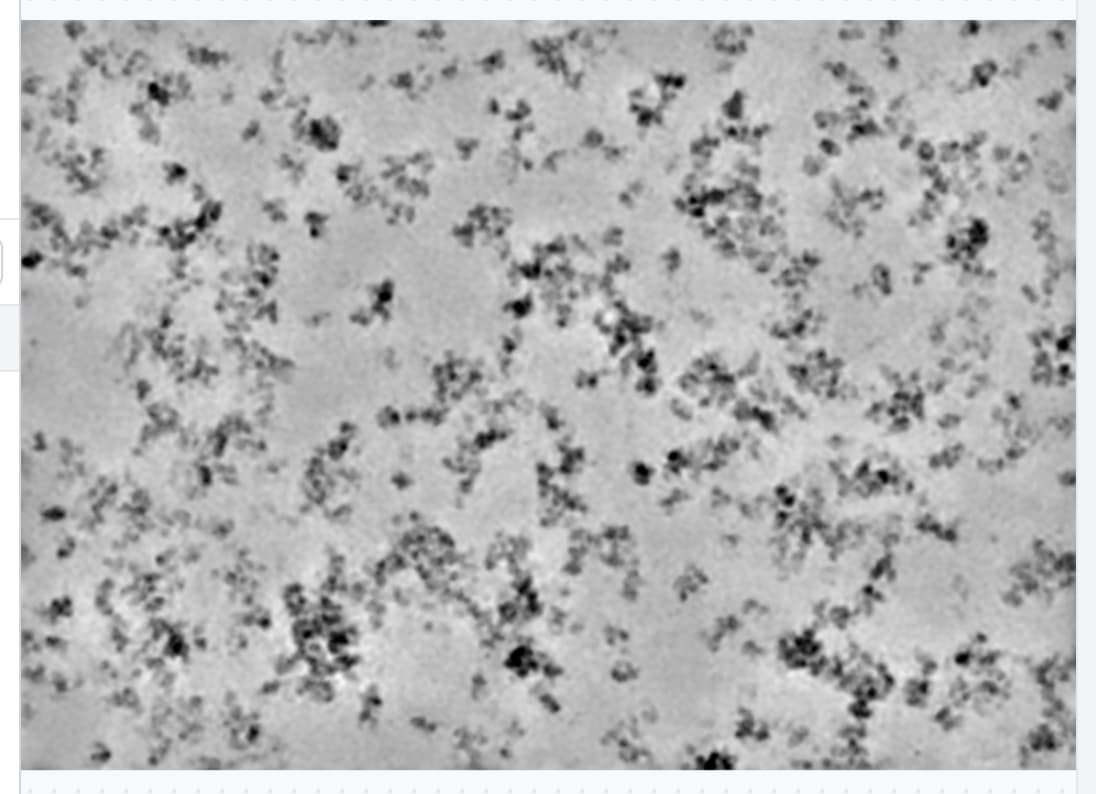
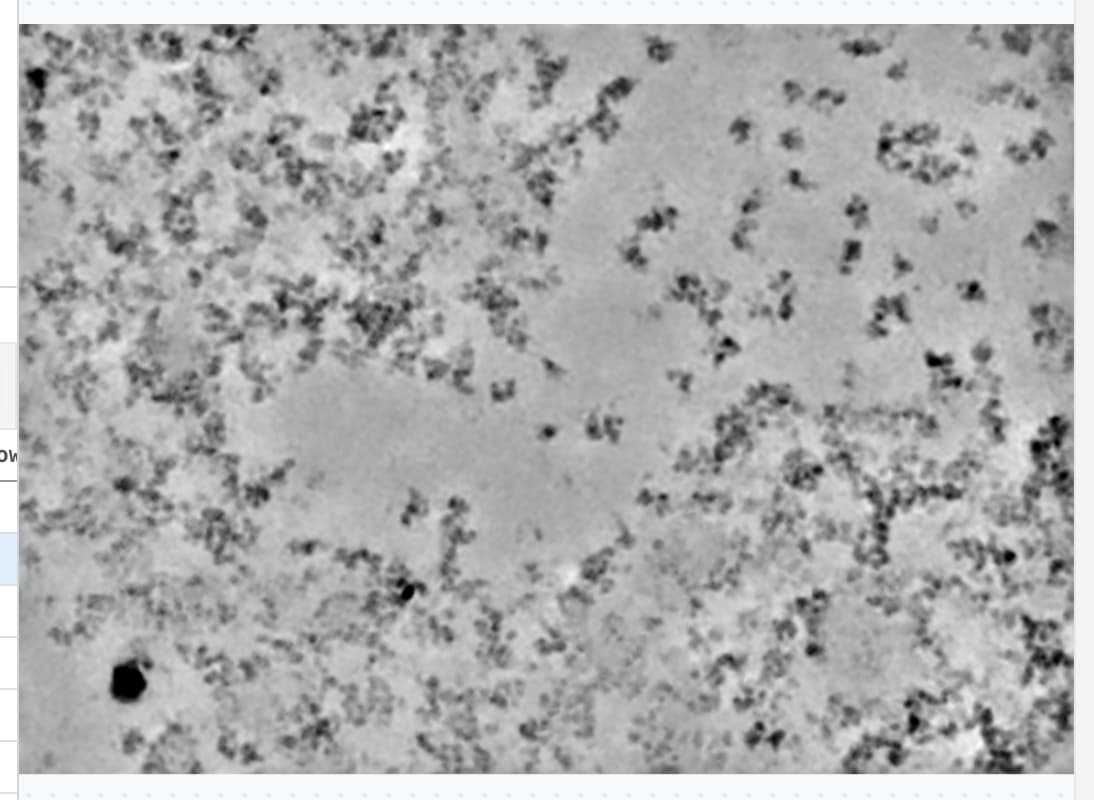
Consider a micrograph like the one attached.
There is a mixture of aggregated (but not denatured) protein with single, clean, particles. This example ultimately gives a nice map at ~3Å, but picking and cleanup are a bit of a pain.
Right now, blob picker (or template picker) do not work optimally for this kind of data - they will pick the clean particles, but also pick all over the aggregate.
Manual picking followed by Topaz works well, but is somewhat laborious.
Given the flat background observed in denoised micrographs, would it be possible to add a variant of blob picker that considers or associates the variance/flatness of the background around the pick center (say in the corners of the extraction box) as a parameter that can be used in particle curation or inspect particle picks?
Or is such a parameter already calculated somewhere during 2D classification, and if so perhaps it could be exposed to the new particle subset tool?
(Alternatively, an enhancement to the junk detector to recognize aggregated protein would also be welcome)
Cheers
Oli