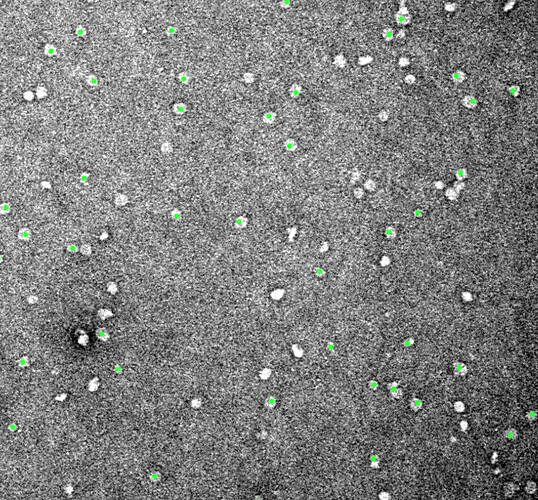
After Inspect picks, it is clearly picking some particles, and it seems to me that the ones it is missing are those with the highest contrast. Perhaps there are some automatic exclusions based on the local power that are being applied behind the scenes, which are appropriate for cryo but not for negative stain? This is with the upper power threshold set to the maximum value: