Hello Colleagues,
We are processing two datasets of the spike protein with different mutations, and we are running into an interesting problem. To pick particles in the first dataset, we downloaded a spike map from the EMDB in the “all-RBD-down” conformation. We low-passed it to 30A and selected a few particles from a few micrographs to generate another template (lets call this template-B). This template-B was used for subsequent particles picking from the entire dataset. We obtained two major conformations (one-RBD-up and two-RBD-up) after 2D classification, ab initio reconstruction, and heterogeneous refinement. All good so far.
Next, we applied the same strategy of using the downloaded EMDB template on the second spike dataset. This gave us the one-RBD-up conformation only. However, when we used template-B on the this second dataset, it shows up both one-RBD-up and two-RBD-up conformations. Can someone please help us understand what is going on and what is the right strategy going forward? Is there some sort of model bias in picking even after a 30A low-pass? Thank you.
I routinely use templates from a closed Spike trimer to pick many different Spike and Spike complex samples, and there’s really no bias in the final result…aside from getting all the expected 3D classes, you can tell that essentially 100% of the particles are picked. Note there will be some difference in the cross-correlation scores, but it won’t be relevant when you are setting thresholds to exclude junk picks.
Is your template B based on 2D classification, or a 3D model? One possible factor could be an incorrect pixel size for the EMDB-based templates in the 2nd dataset.
Also, are your picking parameters really optimized? You should process just 10 micrographs and clear/rerun the picking job varying all of the parameters until it’s as good as can be, usually it takes < 15 tries and about as many minutes. You can also pick Spike almost perfectly using a blob template and optimizing parameters with the same approach (hint - use circle AND ellipse).
Thank you, Daniel. Template-B was generated from 2D classification of our own spike particles. These particles were picked using the EMDB volume imported into cryosparc.
We also ran a test on one single set of micrographs using three different templates of the spike generated from three different EMDB maps: all-RBD-down, one-RBD-up, and two-RBDs-up. Turns out that only ~60% of the good spike particles being picked are common between the three templates. The rest are unique to each template. This is a problem.
You make an interesting point about pixel size. Should I explicitly define the pixel size when importing the EMDB volume? I was under the impression that this would be read from the header. Would an incorrect pixel size cause the problem I mentioned in the above about the lack of common particles from three different templates.
The pixel size should be read from the header, and then dealt with properly when binned according to your output size for Create Templates (I recommend using something like 48-72px as the output, corresponding to about a 6x binning, and then the default 20A filters and angular step are good), but there could be an error somewhere along the line, so good to double check.
How are you using inspect particle picks after the template picking? What diameter, minimum distance, and number of max peaks are you using? Are you picking all the particles?
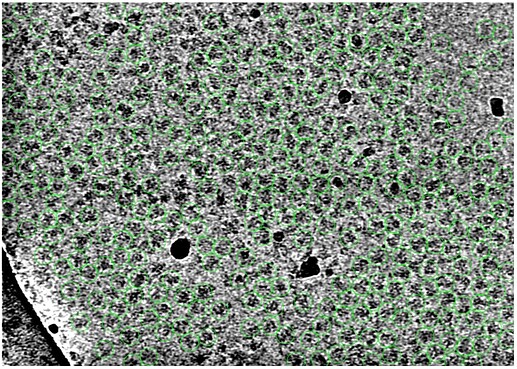
When I say that I pick almost all the particles, this is what I mean:
This is after inspect picks to reject the noise and ice/carbon picks. There are some unpicked particles of course, but mostly because those are clumped up/aggregated with other particles.
Also - if you use RBD open / closed trimer /junk references instead of identical ones, are those classes actually there?
Hello Daniel,
It looks like the pixel information was correctly imported when I imported the volume from EMDB.
Here is a rather dumb question: when a volume is imported from EMDB to generate the template for particle picking, should a pixel size be specified in the import job to re-scale this imported volume to the pixel size in the micrographs? From your comments, I gather that you do not bin the volume during the import job but bin it during the “create template” stage?
Can I ask a second question: how does cryosparc handle discrepancies in the pixel size of the template and micrographs during particle picking? Does it bin/scale the micrographs to the pixel size of the template or does it modify the pixel size of the template? Thank you
The pixel size you can add during Import 3D Volumes will just override the value in the header of the .mrc file. Changing it won’t change the “binning” of the volume - which would require changing the density values in the actual voxels - but it will actually scale the physical extent of the particle relative to the micrographs. Then the size of the templates wouldn’t match the particles. You only need to change this value if the header is has some default value like 1 or the crazy numbers cisTEM puts in there in its intermediate files.
For Create Templates, the projections are interpolated down to that size, so the pixel size increases to keep the physical extent the same (and these values are written into the header of the template .mrc stack). If the the pixel size is 0.8 A/px say, and the template volume was 320x320x320, then choosing a template size of 32 px would bin down to 8 A/px in those 32x32 px template images. In that case the Nyquist frequency is 16 A^-1 so the templates will have slightly higher resolution information than the default lowpass filter of 20 A. They can also have some aliasing, so a slightly bigger box is OK to use, as the higher resolution information will still be ignored due to the low pass filters. (64px ? 72px? So the templates look nice but they are still a small & fast FFT size).
CryoSPARC will use the header values of the .mrc files to scale things appropriately automatically - so as long as those values are right it should work as expected without the user having to do anything.
Thank you, Daniel. This makes sense.
How does your picking compare to the example I showed? I am doing this exact thing all the time (and not just for Spike:Abs, but completely different things like membrane protein complexes), so I’m both pretty sure the bias is avoidable with good (complete) picking, but also concerned / want to make sure we understand what makes it a risk.