I have a suggestion for a new feature in cryoSPARC that is extremely important for symmetry determination of helical samples: an averaged power spectrum. People mistakenly assume that the power spectrum of an average is the same as an averaged power spectrum, and this is not true. The averaged power spectrum is an incoherent average that is invariant under the shifts of images that are needed to generate a coherent real-space average. For example, if one averages together images to look like Albert Einstein, then the power spectrum of this average will look like the power spectrum of Einstein. However, the averaged power spectrum will show no such artifact, and will actually represent the information in the raw images. For helical specimens one may easily see additional information that does not appear in the class average due to heterogeneity. What one needs to do is select the segments associated with the 2D classes desired, impose the rotations on those segments needed to align them to the class average, generate their power spectra, and add these together. Will this be possible?
Regards,
Ed
Dear @egelman,
Thank you for the feature request, and apologies for my delayed response. We will definitely look into generating power spectra using the described procedure, as this would be a great addition to aid helical symmetry determination. It would be fairly straightforward to add this as a standalone post-processing job after 2D classification. I believe another user (@adesfosses) has proposed a similar feature (Proposing a job type to add for helical processing).
In averaging power spectra this way, do you typically try to account for the CTF of the particles? It makes sense that it’s not crucial to account for the CTF when averaging the power spectra compared to the raw images, since only the amplitude of the raw images is used. However, I wonder if this matters when using the power spectra downstream for indexing purposes.
Best,
Michael
Thanks for the response. Indeed, what I have proposed appears to be the same as Desfosses, and he has raised some of the motivation for why this is important. While one needs to do CTF correction to create a 2D class average, the power spectrum does not change if you flip phases or do not flip phases - it is purely the amplitude squared. Of course, one can also introduce amplitude changes when correcting the CTF (such as when one uses CTF-multiplication to improve the SNR) but this will not greatly change the layer lines seen in the averaged power spectrum. Indexing will not depend upon any phase information, since this is completely lost in an averaged power spectrum. If one had a power spectrum from a single filament, then phase information could be extracted. In fact, this was how old Fourier-Bessel processing was done, extracting amplitudes and phases from the FT of each filament. But with extremely weak contrast from thin filaments in ice, this is generally impossible. I hope that this helps.
Regards,
Ed
I have heard nothing more, but wonder if there is simply a mechanism to create a stack file in cryoSPARC of the image segments that have been used in a 3D reconstruction, with the rotations and shifts imposed on them? If such a stack file could be generated, then it would be trivial for us to write a program to generate the averaged power spectrum from such segments. At the moment, the segments have random orientations, and for the average power spectrum we need them to all be aligned to a common orientation of the helical axis. Surely this could not be difficult to implement?
Regards,
Ed
Dear Prof. @egelman,
Currently there isn’t a built-in mechanism to shift/rotate and re-write out a particle stack in cryoSPARC. Unfortunately the closest thing to that is 2D classification, but this doesn’t address the issue. We are looking to add this feature as soon as possible, but averaging images’ power spectra would require somewhat nontrivial additions to our engine, so it would likely have to wait until the next release.
In the meantime it may be possible to accomplish this via scripting, but would likely be laborious. The alignments2Dare present in any .cs file of 2D-classified particles, which contains the axis-angle rotations specifying the in-plane angle of the particles in units of radians. It may be possible to write out a stack of aligned particles using a .cs file of particles from 2D classification in conjunction with an external software or python package capable of image rotation, such as pyem. However, I can’t advise more as I do not have much experience using pyem.
Best,
Michael
Thanks. We do not need the averaging of power spectra (we can do that quite easily ourselves), only the aligned stack itself. The .cs file appears to be binary, so that will not be useful.
Regards,
Ed
@egelman The .cs files are just serialized numpy arrays; you can read it with numpy.load().
Actually, I think you should be able to export them with csparc2star.py and then use relion_image_handler to apply the alignments. (--avg_ampl2_ali?).
Piggybacking off of @DanielAsarnow’s comment: you can find a tutorial written on our guide about manipulating .cs files in python (the example is using scripting to re-center particles).
Best,
Michael
There is now a feature in cryoSPARC to generate an averaged power spectrum from segments belonging to a particular 2D class average, but there are two (related) problems with it:
- it is slow
- it tries to actually rotationally align the individual power spectra, which is why it is slow
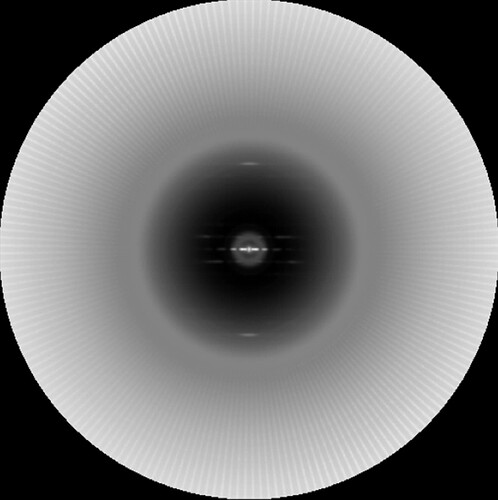
To show why (2) is problematic, here is such an averaged power spectrum generated by cryoSPARC:
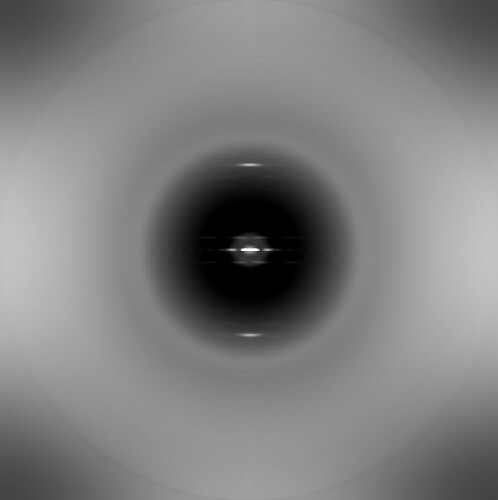
If we generate an averaged power spectrum using SPIDER, with these same segments, there is no such artifact (the radial lines at high resolution):
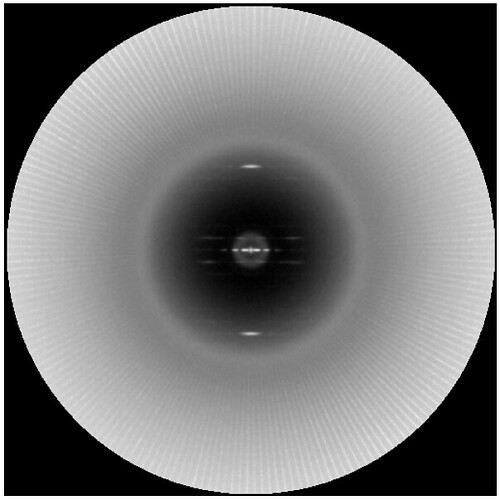
The artifact arises from the fact that the first frame of the movies has a gain normalization problem so that columns of pixels have different mean intensities. This can be seen taking a power spectrum of a single image:
In the Spider averaged power spectrum, these lines would be spread out over all orientations (due to the fact that the segments will have random orientations with respect to the camera raster). But in cryoSPARC, these lines are being aligned into discrete orientations.
There is no need to do such alignments of the power spectra in cryoSPARC. For every 2D class average, the orientation of every segment is known. One needs to simply rotate each segment by those parameters, generate a power spectrum from each, and add them together. That would be very fast, as the “heavy lifting” (finding the orientation of every segment) has already been done. Not only is the present approach unnecessarily slow, but it can introduce artifacts such as the one I just showed.
Dear @egelman,
Thanks for your detailed post – this is quite an interesting case comparison between SPIDER and CryoSPARC. The main reason why the power spectra averaging in CryoSPARC is fairly slow relative to our other jobs is that performance wasn’t prioritized during the implementation, and it is not GPU accelerated. The job itself simply reads the alignments2D from the input particles, rotates them accordingly, and averages their power spectra together; it doesn’t attempt to estimate any rotational alignment of the segments or the power spectra.
The comparison is interesting; if I may confirm a few questions:
- Is the gain normalization issue present in all movie frames? If so, does the gain reference appear to account for for this “streak”?
- If the gain reference doesn’t correct the “streak”, is it possible to correct the gain normalization issue via creating a rough gain reference by manually averaging together many movie frames in the dataset?
- Are the raw particles (segments) identical across both the SPIDER and CryoSPARC images you shared, or was particle extraction (and/or motion correction) done separately in each software? If done separately, does the gain normalization issue in SPIDER appear resolved?
- Are the 2D alignments the same across SPIDER and CryoSPARC?
Unfortunately the quality of 2D alignments may be significantly impacted by a stark normalization issue such as the one present, so it may be worth repeating 2D classification & power spectrum averaging after correcting for the normalization issue. The appearance of the radial streaks in CryoSPARC’s averaged power spectra may be indicative of the fineness of our 2D rotational search, which is about ~1º after all branch and bound iterations.
Best,
Michael
Thanks for the very full response. The gain normalization issue is an artifact of the first movie frame. So if one creates a dose-weighted motion corrected image, then this will be there unless the first frame is excluded. Then this issue disappears. Since you say that alignment of power spectra is not being done, an alternative possibility is that in the 2D average discrete orientations are being chosen based upon aligning “noise”. So imagine that a rough low-resolution alignment is achieved with a segment, but rotating it by +/- 3 degrees aligns this noise. Is that possible? There are several things that we cannot understand. The power spectrum above was generated with ~ 200k segments. If we used 14k segments, we get a finer pattern of radial streaks:
However, when we used 3k segments it was coarser, and it is hard to reconcile this with 1° increments in the 2D rotational search:
To answer the other questions, the particles used for Spider were identical to those used by cryoSPARC, and were simply exported as .spi files. In Spider, we aligned the particles to the cryoSPARC class average. This alignment is the slow step in Spider, while the fast step is simply imposing these alignment parameters. Nevertheless, determining the alignment, imposing the alignment and generating the power spectra were faster in Spider than just imposing the alignment and generating the power spectra in cryoSPARC. Needless to say, there is no GPU acceleration in Spider.
It is possible that the radial streaks in the power spectrum of each segment, if the first frame is included, might degrade the 2D class averages due to the possibility that these streaks are being aligned. But removing the first frame, and this artifact, leads to no improvement in the 2D averages.