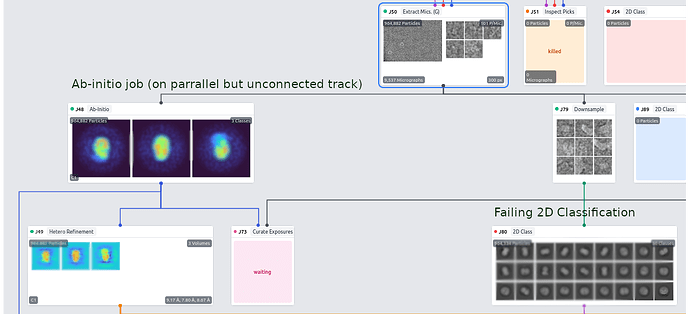
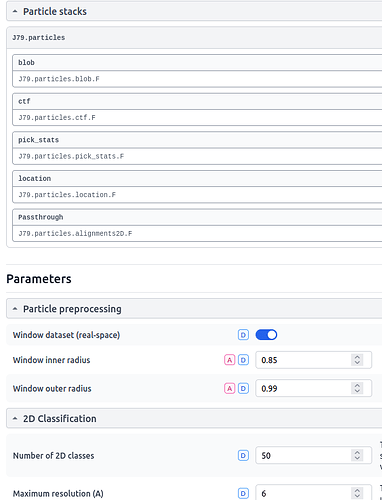
I’m attempting 2D classification and this error is reported near the conclusion of the job (when compiling the passthrough results for the particle stacks):
[CPU: 5.70 GB Avail: 237.25 GB] Passthrough results ['pick_stats', 'location', 'alignments_class_0', 'alignments_class_1', 'alignments_class_2']
[CPU: 5.70 GB Avail: 237.25 GB] Traceback (most recent call last):
File "cryosparc_master/cryosparc_compute/run.py", line 100, in cryosparc_compute.run.main
File "/home/bio21em2/Software/Cryosparc/cryosparc_worker/cryosparc_compute/jobs/runcommon.py", line 1074, in passthrough_outputs
dset = load_input_group(input_group_name, passthrough_result_names, allow_passthrough=True, memoize=True)
File "/home/bio21em2/Software/Cryosparc/cryosparc_worker/cryosparc_compute/jobs/runcommon.py", line 664, in load_input_group
dsets = [load_input_connection_slots(input_group_name, keep_slot_names, idx, allow_passthrough=allow_passthrough, memoize=memoize) for idx in range(num_connections)]
File "/home/bio21em2/Software/Cryosparc/cryosparc_worker/cryosparc_compute/jobs/runcommon.py", line 664, in <listcomp>
dsets = [load_input_connection_slots(input_group_name, keep_slot_names, idx, allow_passthrough=allow_passthrough, memoize=memoize) for idx in range(num_connections)]
File "/home/bio21em2/Software/Cryosparc/cryosparc_worker/cryosparc_compute/jobs/runcommon.py", line 635, in load_input_connection_slots
dsets = [load_input_connection_single_slot(input_group_name, slot_name, connection_idx, allow_passthrough=allow_passthrough, memoize=memoize) for slot_name in slot_names]
File "/home/bio21em2/Software/Cryosparc/cryosparc_worker/cryosparc_compute/jobs/runcommon.py", line 635, in <listcomp>
dsets = [load_input_connection_single_slot(input_group_name, slot_name, connection_idx, allow_passthrough=allow_passthrough, memoize=memoize) for slot_name in slot_names]
File "/home/bio21em2/Software/Cryosparc/cryosparc_worker/cryosparc_compute/jobs/runcommon.py", line 625, in load_input_connection_single_slot
output_result = com.query(otherjob['output_results'], lambda r : r['group_name'] == slotconnection['group_name'] and r['name'] == slotconnection['result_name'], error='No match for %s.%s in job %s' % (slotconnection['group_name'], slotconnection['result_name'], job['uid']))
File "/home/bio21em2/Software/Cryosparc/cryosparc_worker/cryosparc_compute/jobs/common.py", line 683, in query
assert res != default, error
AssertionError: No match for particles.alignments_class_0 in job J80
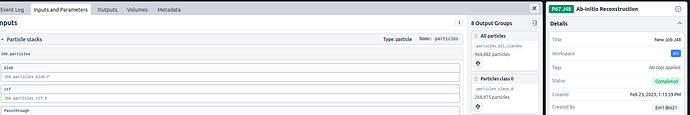
There 3D class alignments should not be part of the particle stacks, all that has been done is an extract and then a downsample job. The particle stack seems to be picking up pass-through parameters from an ab-initio job (J48) that was performed on a parallel but unconnected processing pathway and then J80, the 2D classification, fails when this information is not found in J79, which is the parent job:
CryoSPARC instance information
- Type: master-worker
- Current cryoSPARC version: v4.1.2
- Linux Freyja 5.4.0-89-generic #100-Ubuntu SMP Fri Sep 24 14:50:10 UTC 2021 x86_64 x86_64 x86_64 GNU/Linux
total used free shared buff/cache available
Mem: 187 18 0 0 168 167
Swap: 1 1 0
on master node
CryoSPARC worker environment
$ env | grep PATH
CRYOSPARC_PATH=/home/bio21em2/Software/Cryosparc/cryosparc_worker/bin
PYTHONPATH=/home/bio21em2/Software/Cryosparc/cryosparc_worker
CRYOSPARC_CUDA_PATH=/usr/lib/nvidia-cuda-toolkit
LD_LIBRARY_PATH=/usr/lib/nvidia-cuda-toolkit/lib64:/home/bio21em2/Software/Cryosparc/cryosparc_worker/deps/external/cudnn/lib
PATH=/usr/lib/nvidia-cuda-toolkit/bin:/home/bio21em2/Software/Cryosparc/cryosparc_worker/bin:/home/bio21em2/Software/Cryosparc/cryosparc_worker/deps/anaconda/envs/cryosparc_worker_env/bin:/home/bio21em2/Software/Cryosparc/cryosparc_worker/deps/anaconda/condabin:/home/bio21em1/Software/ResMap:/home/bio21em1/Software/CTFFIND/bin:/usr/local/IMOD/bin:/home/bio21em1/Software/relion/build/bin:/home/bio21em1/Software/cryosparc3/cryosparc_master/bin:/home/bio21em1/bin:/home/bio21em1/anaconda3/bin:/home/bio21em1/anaconda3/condabin:/usr/local/IMOD/bin:/usr/local/sbin:/usr/local/bin:/usr/sbin:/usr/bin:/sbin:/bin:/usr/games:/usr/local/games:/snap/bin
$ which nvcc
/usr/lib/nvidia-cuda-toolkit/bin/nvcc
$ nvcc --version
nvcc: NVIDIA (R) Cuda compiler driver
Copyright (c) 2005-2019 NVIDIA Corporation
Built on Sun_Jul_28_19:07:16_PDT_2019
Cuda compilation tools, release 10.1, V10.1.243
$ python -c "import pycuda.driver; print(pycuda.driver.get_version())"
(11, 7, 0)
$ uname -a
Linux Odin 5.4.0-99-generic #112-Ubuntu SMP Thu Feb 3 13:50:55 UTC 2022 x86_64 x86_64 x86_64 GNU/Linux
$ free -g
total used free shared buff/cache available
Mem: 251 12 7 0 231 237
Swap: 231 5 225
$ nvidia-smi
Mon Feb 27 15:07:05 2023
+-----------------------------------------------------------------------------+
| NVIDIA-SMI 510.73.05 Driver Version: 510.73.05 CUDA Version: 11.6 |
|-------------------------------+----------------------+----------------------+
| GPU Name Persistence-M| Bus-Id Disp.A | Volatile Uncorr. ECC |
| Fan Temp Perf Pwr:Usage/Cap| Memory-Usage | GPU-Util Compute M. |
| | | MIG M. |
|===============================+======================+======================|
| 0 NVIDIA GeForce ... Off | 00000000:18:00.0 Off | N/A |
| 27% 23C P8 9W / 250W | 299MiB / 11264MiB | 0% Default |
| | | N/A |
+-------------------------------+----------------------+----------------------+
| 1 NVIDIA GeForce ... Off | 00000000:3B:00.0 Off | N/A |
| 26% 24C P8 8W / 250W | 160MiB / 11264MiB | 0% Default |
| | | N/A |
+-------------------------------+----------------------+----------------------+
| 2 NVIDIA GeForce ... Off | 00000000:86:00.0 Off | N/A |
| 26% 25C P8 8W / 250W | 160MiB / 11264MiB | 0% Default |
| | | N/A |
+-------------------------------+----------------------+----------------------+