Why does the denoiser require movies?
Formally, the denoiser requires half micrographs, but these can only be generated by producing separate micrographs from even and odd frames of movies.
The half micrographs are required because the denoiser model is trained using two noisy estimates of the same data. As explained in the original Noise2Noise paper (by which the denoiser is heavily inspired), having two estimates of the same data with independent noise makes it possible to model the underlying noiseless images.
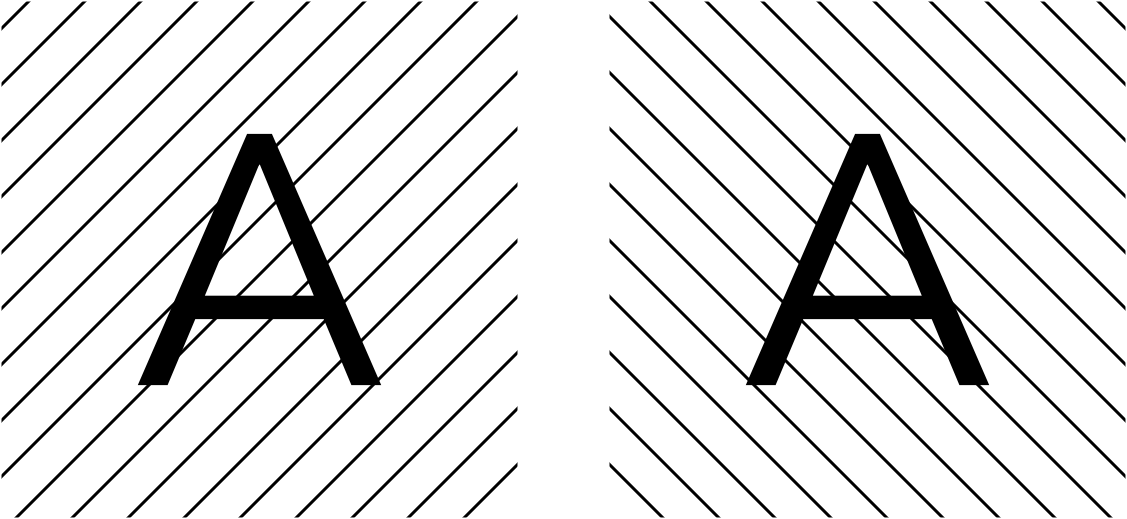
For an intuitive example, consider these two “half-images” which I’m telling you look identical, except for noise:
Since you know that, in the noiseless case, these images should be identical, you know anything that differs between the two half-images is noise (there’s an analogy to be made here with the GSFSC as well). Knowing this, you can produce a denoised image (ideally, just the letter A on a white field). This is the underlying principle of Noise2Noise and the Micrograph Denoiser.
Let me know if it’s still not clear why training a new model requires movies/half-mics! I’m still figuring out the best way to explain it 
How much does the quality of the training data impact performance?
I haven’t done enough extensive testing to give you a quantitative answer here, unfortunately. In my experience, it’s rather robust with regard to the number of training micrographs and the amount of those micrographs covered by ethane contaminants, ice crystals, hole edges, etc. If the grayscale estimate is bad, that does hurt performance.
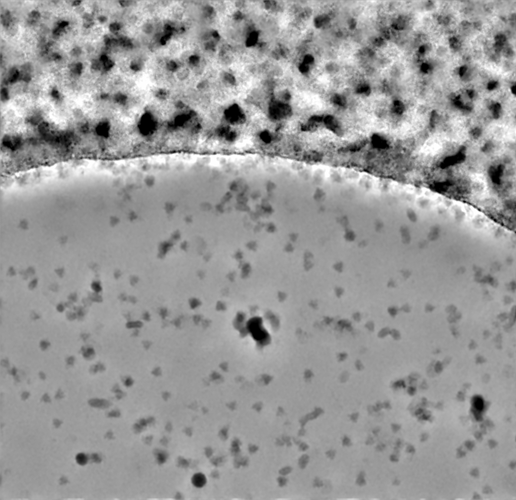
I quickly pulled 16 movies from EMPIAR 11632 (chosen just because I had it to hand) which had a lot of carbon or ice and trained a denoiser using just those 16 movies with otherwise all default settings. I think it did pretty well, considering:
Indeed, if you have or find any examples where training fails to produce a good denoiser, I’d be interested in seeing them!
Feature request
I’ve recorded your request for adding the Denoiser to CryoSPARC Live!