Hello everyone,
I’m working with a dataset collected on the Krios microscope for a membrane protein, and I’ve encountered an issue that I don’t fully understand.
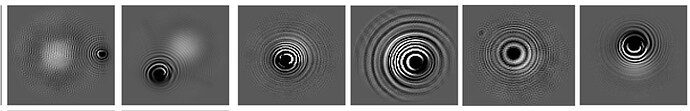
In both my micrographs and 2D classifications, I’m seeing random rings appear with varying frequencies (as shown in the attached image).
I’ve tried several methods to remove particles affected by these rings, including multiple rounds of 2D classifications and heterogeneous refinement, etc. However, these strategies still result in a map with very strong anisotropy. It seems that some of the rings, especially those with very low frequencies, might be contributing to this problem.
Has anyone experienced a similar issue or have any insight into the possible causes of these rings? I’d also greatly appreciate any suggestions on how to reduce the anisotropy in my final maps.
Thank you in advance for your help.