Hi,
I find the reference matching performed in Reference Auto Select 2D very useful, but it can be sometimes difficult to find exactly the right parameters to automatically select the correct classes.
The visual comparison of the corresponding reference projections is very handy though - would it be possible to either add an interactive step where the user could edit the selection, or incorporate matched reference projections to sort and visually compare the classes in “regular” select 2D?
2 Likes
I think I misunderstood the point. Could you do the same sort of thing with multiple runs with manually selected references (see below)?
Old post for posterity:
Involves extra steps, but you can select templates only with Select 2D, so could you create projections, select the views you want and then feed that to the Reference Auto Select? I’ve not used it (yet) so not sure of what options there are.
No - the way it works is you supply a volume, and it matches projections of that volume to 2D class averages (and associated particle sets)… so select 2D wouldn’t help in this instance I think?
Ah, yes; I see there is no option to provide 2D projections yourself, just a 3D. Sorry, I’ve used the select 2D trick a few times recently to specifically go hunting for rarer/low contrast views when picking so wondered whether it would also apply here.
That seems like a wasted opportunity, as I’d see reference select 2D as a good chance to go hunting specifically for rarer views…
1 Like
It also make sense to visually access the results - in my tests the empty / blank classes can we considered as acceptable and selected during Reference based Auto Select 2D.
Kind regards, Dmitry
I quite like it also for compositional heterogeneity - e.g. for a nanobody complex, getting rid of 2d classes that lack the nanobody prior to ab initio. Sometimes can be easier to tell when you see the matched projections!
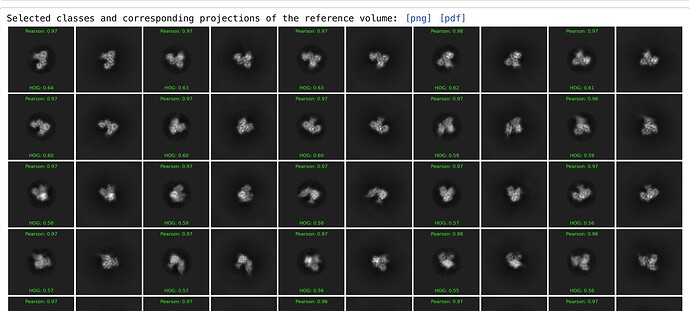
Here is an example that perhaps illustrates the point better - comparing projections of 1ATP (crystal structure, molmapped at 6Å) to 2D classes of EMPIAR-10252 (same molecule, cryo dataset):
The comparison with the projections is very helpful, visually, and allows identification of some classes that might be otherwise discarded. But clearly some of the pairings are bogus, so it would be helpful to be able to manually edit/interact with the outputs.
I would love to see this feature as well. I use reference based select 2D to help train my eyes to see which classes are actually my protein, and which are just junk. What I end up doing is just running this job to see which classes I should choose, and then visually comparing it Select 2D to pick the classes to move forward with.
In the absence of making reference based select 2D interactive, it would be helpful to label the output of this job with the same class numbers as 2D classification sorted by by number of particles in descending order.
Hi all - thanks for your feedback! We’ve recorded this request.
1 Like