I am trying to do 2D class for a small protein complex (~90KDa). I turned off "Force Max over poses/shifts " hoping to obtain better 2D classes, but ended up getting featureless dot for all the classes. I’ve never used this feature before, so I don’t know what could cause this. Can anyone help me with this, please?
Hi, I’m not sure what is causing this, but I do notice that your box looks very large with respect to the particle size. Maybe try extracting in a smaller box, maybe half that size?
Cheers
Oli
Hi @Meiling, (and thanks @olibclarke!) - could you also let us know whether you were seeing reasonable classes before turning off Force Max ?
Thank you, @apunjani and @olibclarke for replying to my question.
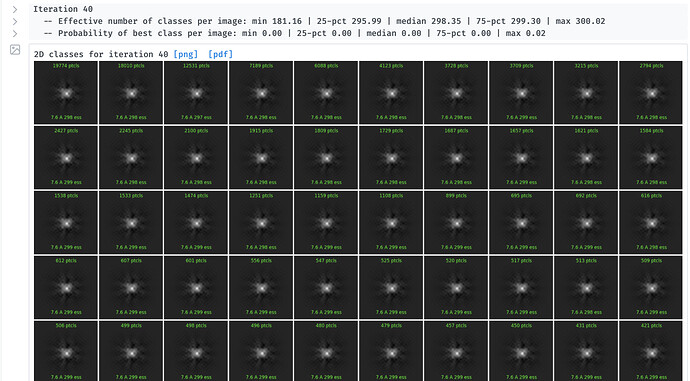
Yes, I saw classes before I turned off Force Max (pic attached) although the background was noisy. I was trying different things hoping to get better classes and ran into the described issue with Force Max.
I did try setting the box size to 150 A (my particle diameter is about 110 A), and it gave me basically the same result (featureless bright dot for all classes).
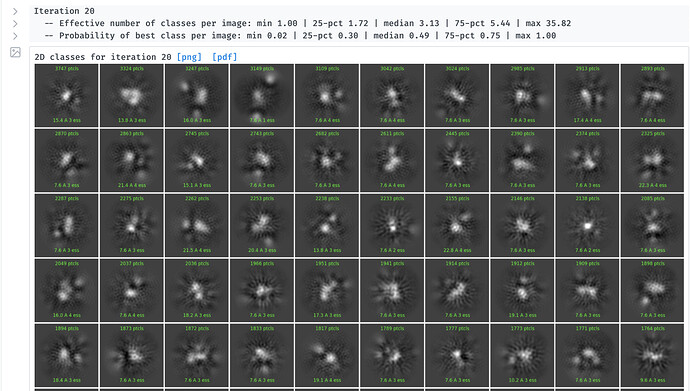
By the way, I also tried turning on “Clamp-solvent” and “Enforce non-negativity” (while kept “Force Max” on), and seeing classes like below. I’m wondering how reliable the clamp-solvent model is and would it affect the subsequent analysis? I’m seeing lots of details in the classes, I wonder how reliable these classes are. I’m new to this, so I would appreciate any feedback. Thank you again!
Hi @Meiling,
The clamp-solvent method (with enforce non-neg on) runs a sub-optimization problem for each 2D class in order to construct a non-negative valued 2D reference that best matches the aligned particle images. In some cases, due to the enforcement of the strong non-negativity constraint, the classes can substantially improve in their quality and detail. But in some cases, the constraint creates only the impression of higher resolution detail… we’ve experimented with it a fair but but have not found a conclusive strategy or way to tell when to use it, so we keep it off by default. Usually the litmus test is if you see multiple classes with distinctive and recognizable features that are clearly the same parts of the molecule. If you see what are clearly different views (even slightly different) that is also a trustable signal. But in your case I wouldn’t feel comfortable basing much analysis on these 2D class results.
That said, it may be worth simply jumping over to 3D and start doing 3D ab-initio reconstructions (optionally with multiple classes), to see what comes out. Sometimes with all particles in 3D, things work better than in 2D.
Also, for sanity checking, could you post an image of one of your micrographs (maybe low-pass filtered from the inspect picks job)? It would serve as a good way to tell what to expect processing to look like, so we can say whether maybe something else is going wrong (ctf estimation etc)
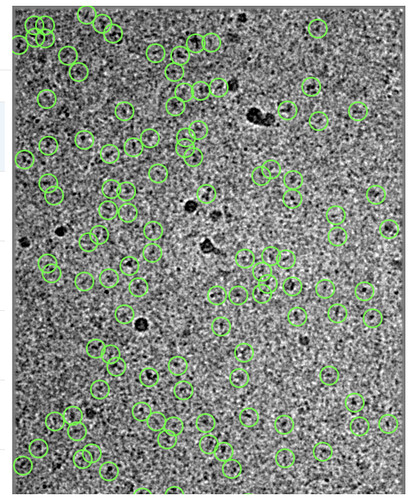
Thank you, @apunjani ! I’m attaching one of the micrographs (with 20 A low-pass filtered). The particles I’m looking for are about 1105060 A, and in some views, the particles bend in the middle. What do you think of the micrograph? Thank you again!
Hi @Meiling,
Sorry for the delayed response.
In this case, I would be slightly suspect of the data… there appear to either be many, many particles that are not being picked, or else many of the particles that are being picked are not true particles…
Have you already done some negative stain or other experiments that verify the overall size and shape of the particle? Do you have any micrographs collected at higher defocus where low-resolution contrast is high enough that you can tell what the particles definitely look like?
Perhaps others on this forum will have some input!