Hi guys! I’m quite new to cryoEM, and I’m playing with CTF and cryoSPARC Simulate Data (BETA) job, but I find that results of my own CTF code are quite different from the ones cryoSPARC Simulate Data (BETA) job gives. As I can only post one figure, I combine all figures in one and insert it at the end.
Here is the thing. I want to get some clean projections of a volume first (this can be done by cryoSPARC or other softwares like RELION, EMAN2, etc), and then add CTF effect with given parameters to these projections. To my understanding, this can be done by taking the Fourier transform of a projection, multiplying the 2D CTF image in Fourier space, and doing the inverse Fourier transform.
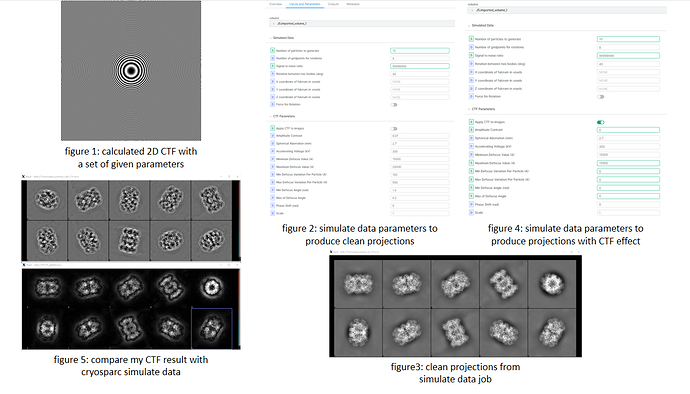
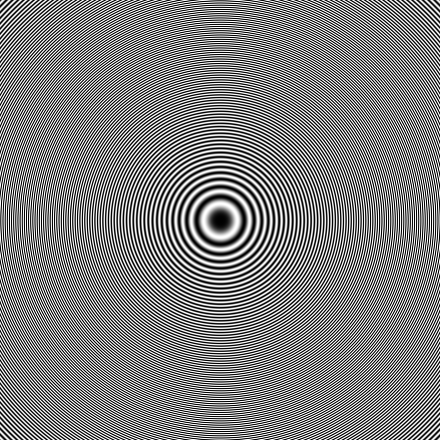
So, I calculate the 2D CTF image according to the CTF formulation. ( [CTF Estimation | CryoSPARC Guide]) It looks like figure 1 with parameters: defocus=15000 angstrom, voltage=300kV, Cs=2.7mm, phase shift=0rad, pixel size=0.6575, image size=440. The first 4 parameters are sufficient to calculate CTF value according to the formula on the website, and the last 2 parameters are consistent with the imported volume.
Clean Projections of the volume are produced by cryoSPARC Simulate Data (BETA) job. I set the SNR to a really really big value, and turn off the CTF option. The overall parameters are shown in figure 2. I think the result particles are very close to clean projections of the imported volume, as shown in figure 3. (It’s viewed with EMAN2 e2display.py)
To add CTF, the .mrcs file is read by mrcfile python package firstly, and CTF effect is added to the projections by numpy as:
mrc = mrcfile.open(file_name, mode=‘r’)
data = mrc.data
mrc.close()
proj = data[k, :, :] # shape (440, 440), k iterates over all projections
ctf_2D = … # ctf_2D is the one shown in figure 1
proj_with_CTF = np.abs(np.fft.ifft2(np.fft.fft2(proj)*np.fft.fftshift(ctf_2D)))
CTF_added_proj_stack[k, :, :] = proj_with_CTF
… # write .mrcs file with CTF_added_proj_stack
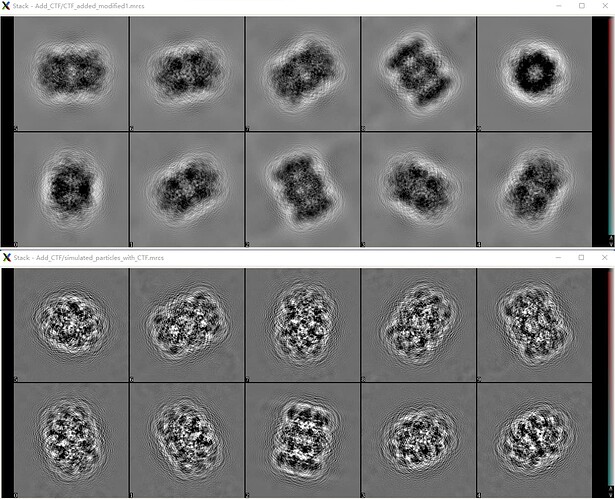
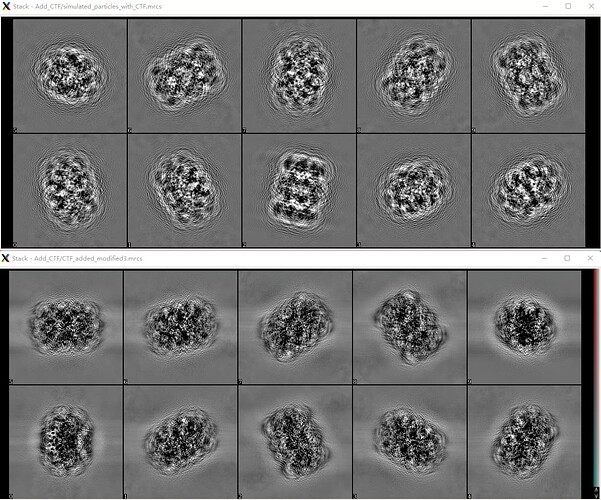
When I turn on the CTF option in Simulate Data (BETA) job, set the CTF parameters the same as used in my own code, and remain the noise parameters the same, I also get the projections affected by CTF. (Overall parameters are shown in figure 4, and I think it matches the parameters used in my code, which is described above to produce figure 1) As shown in figure 5, however, comparing the results of my code (figure 5 bottom) and cryoSPARC Simulate Data (BETA) (figure 5 top), there exists significant difference. (viewed in EMAN2 e2display.py)
I have tried but can’t figure out what’s wrong. I also tried to use projections produced by EMAN2 and add CTF effect by my code, but the results are similar as described above.
Why is it? Do I make mistakes on data processing or CTF calculation? Or do I miss some steps to add CTF effect? Anyone can help me? Thanks for any suggestions!
Cheers
Ciren