Hi,
This is a very stupid question.
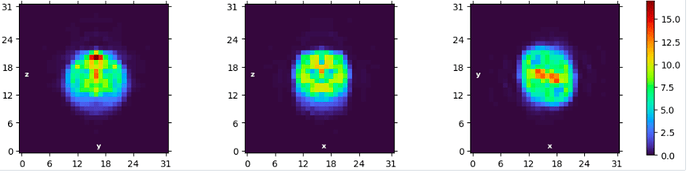
I collected a dataset using arctica with 0.51 apix, at 165k mag. But when processing through ab-initio I started getting classes with broken pixels (image attached).
It feels like I messed up something but not sure.
Please advise.
Thanks
I’m skeptical that you’re looking at a 10 angstrom particle. How large is the particle expected to be? Could you just be off by a factor of 10 in your import? Or maybe box size somewhere and it’s picking up noise?
I’m not entirely sure about the diameter of the particle but it’s a 70kD protein in nanodisc. I could see the particles clearly that’s why I didn’t consider to be junk. I started using 400 pix box size and downsampled to 200 pix. I never changed any parameter except default CS params during import.
your pixels are not broken - it’s just that the box size is set according to the desired final resolution to use for ab initio (default 12 Å). This speeds computation. In your case, that means a box of 32px.
If you want to get the same reconstruction in your original box, you could take the output of ab initio and perform a homogeneous reconstruction job.
Alternatively, you may want to play with the ab initio parameters anyway - I often find that for small proteins in nanodisc or detergent, starting at higher resolution works better. Often starting at 9Å and going to 7 or even 5 Å final works well.
It also looks like you have enforced symmetry (C2?) during ab initio. This can be useful in certain cases, but I wouldn’t recommend it for a first attempt.
Cheers
Oli
1 Like
Thank you so much @olibclarke for the detailed explanation. This is my first time with this small particles and with this high mag. So I was little skeptical.
Yes, I did use C2 to get a faster reconstruction but with many noises I see your point.
Thanks for clarifying in detail.
Update: This strategy helped. Can see the transmembrane regions now, however, awaiting for better resolution.
Thanks again.