Hi,
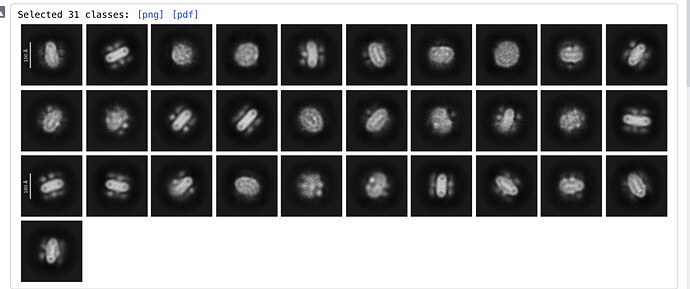
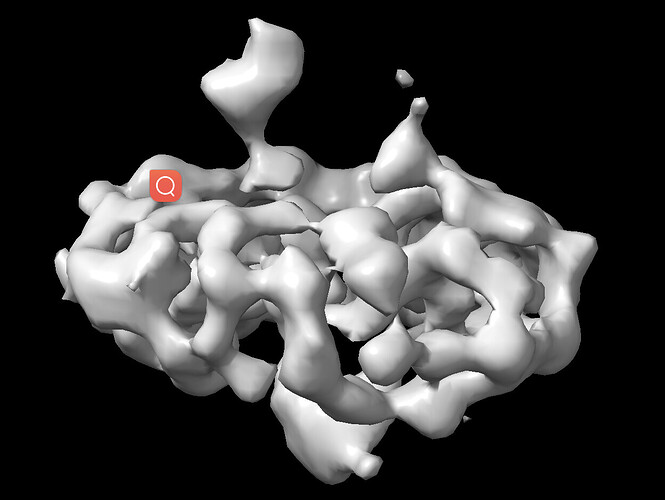
My protein is a multiple-copy oligomer in the membrane. The cytosolic region is small (~7 kDa). After three rounds of 2D classification, I obtained 34,000 particles (see the attached image). I selected ~3,500 particles for ab initio reconstruction, hoping to generate a reference model. However, the resulting model appears flattened, and it is difficult to discern any helical features. I would appreciate any suggestions or insights on how to improve the process.
Ab initio won’t give you the visibility of helices.make sure that do not put high resolution cutoff. A high 8 and low 15 A should work fine. Your 2d seems to give you the expected features. However i assume that you 3,500 particles are too less to give you a meaningful volume according to your expectation.
I would increase particles for a reference volume. And also remember the fact that the 7kDa membrane protrusions wont be very much noticeable from your micelles.
A different approach can be; After you generate a volume, add that as well as the junk ones to a iterative heterogeneous refinement and add all the input particles of 2D classification(removing obvious junks) to sort them out followed by a Nu refinement.
Hope that works.
Thanks for your advice.I will try.