Dear cryosparc community,
You paper on 3DVA is very interesting. I was trying to replicate what you did with big ribosomal subunit ensemble with my data.
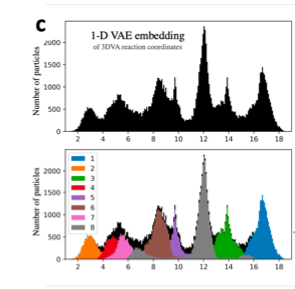
Is there a way to use the clustering method of 1-D VAE embedding with my data?
Thank you for your attention
Best regards,
Nuno Bustorff
1 Like
It’s been described here: Output particles in "intermediates" mode of 3D-variability display?
The GMM is better for this example, probably, because you can clearly count all of the modes.
Thank you @DanielAsarnow.
I will read the thread carefully.
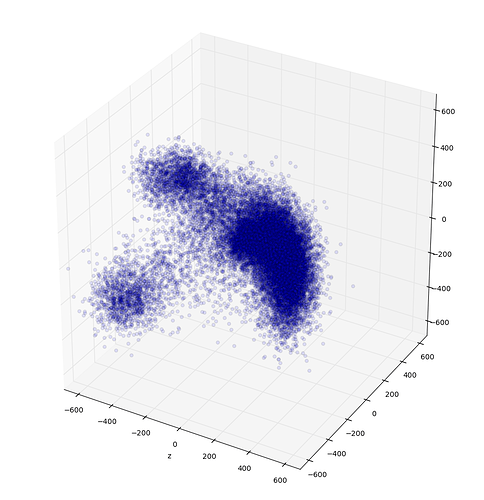
Just for your information this is how my output of 3DVA looks like.
I clearly see three clusters but I don’t know if the data has more sub clusters, which from the 1-D VAE I thought that should give me a better perspective and allow me to get all particles that belong together for further refinement.
Thank for your help once again
Best regards,
Nuno
Looks cool - I think in your case 3DVA is working well and there is some real discrete heterogeneity that can be resolved. Seems like there are the two clearly separated Gaussian clusters, plus the crescent shaped one which could be profitably subdivided (or not). I would try a few different numbers of Gaussians in the GMM so you can get 1 for each satellite cluster and a few that capture the crescent well (maybe 3-4 that can then be grouped together). Then see what the satellites and crescent each represent, by doing new refinements and looking at the 3DVA display results. This way is nice because you can do it all in cryoSPARC easily.
The 1D VAE or other further non-linear embedding might separate them well too, but I’ve found they often still overlap a lot. Manual “gating” of the values could be easier.
Thanks for the advices! @DanielAsarnow