Hi,
I currently created custom mesh for 3D flex mesh preparation job that has 3 meshes in total.
However, when I download the output mesh.pdb file and open it on chimera X or pymol to check, I can only see one mesh. That one mesh looks like sum of the all 3 meshes but I would like to be able to see individual sub meshes to check if they were created correctly.
Is there a way to visualize individual sub meshes, instead of a one combined mesh? or have i done something wrong and lost sub meshes?
Thanks in advnace
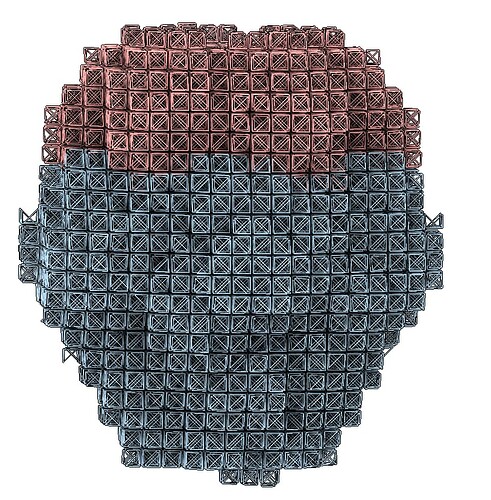
Hi @Jecy! Right now, the PDB visualization only ever shows a single mesh, regardless of how many submeshes there are. So your result is probably correct, but hard to interpret in the PDB mesh. You can check the output of the Mesh Prep job. If your overall mesh PDB looks right, and you see different colors in the “segmented mesh” plot (like below), the job mostly likely did what you wanted.
If you want to visually inspect the meshes in more detail, I have written a python script which creates a BILD file you can view in ChimeraX which shows all of the meshes and vertices, below. You will need to install polars, pandas, and cryosparc-tools for this script to work. If you need help running this script, you can find more information about cryosparc-tools here and here. I will warn you, it’s a bit slow, since I didn’t expect to share it and haven’t optimized it at all. Also, the BILD files are quite large, so they take a while for ChimeraX to draw them.
Running the script produces a set of BILD files. The BILD files ending in g{n} is the tetra mesh for group N. The file ending in verts is the vertices, along with annotations about which groups that vert is used in, and whether those groups are fused. The combined file has everything in a single BILD file.
You can change the color of the mesh groups if you like using the --color-scale option. For example, here is the result of running ./mesh_to_bild.py P345 J749 --color-scale Pastel1
I have found that some meshes are easier to inspect with an orthographic camera (camera ortho).
I hope that’s helpful!
mesh_to_bild.py
#!/usr/bin/env python
from cryosparc.tools import CryoSPARC
import json
import numpy as np
import polars as pl
import pickle
from itertools import combinations
from pathlib import Path
import matplotlib.colors
import argparse
import shutil
def group_color(plot_colors, group_number):
"""
Select a color, looping around the list instead of failing
"""
return plot_colors[group_number % len(plot_colors)]
def random_three_vector():
"""
Generates a random 3D unit vector (direction) with a uniform spherical distribution
From http://stackoverflow.com/questions/5408276/python-uniform-spherical-distribution
"""
phi = np.random.uniform(0, np.pi * 2)
costheta = np.random.uniform(-1, 1)
theta = np.arccos(costheta)
x = np.sin(theta) * np.cos(phi)
y = np.sin(theta) * np.sin(phi)
z = np.cos(theta)
return np.array((x, y, z))
def write_multi(file_obj_list, line_to_write):
"""
Helper function to write the same string to multiple files
"""
for f in file_obj_list:
f.write(line_to_write)
def paste_vectors(group, src, dest):
"""
Create a string representation of a single tetra edge, per group.
"""
group = str(group)
src = ",".join(str(x) for x in src)
dest = ",".join(str(x) for x in dest)
return f"{group};{src};{dest};"
def shrink_tetra(verts, scaling_factor):
"""
Shrink a tetra's verts toward its center of mass by scaling factor. Or I guess its inverse.
1.0 keeps the tetra at full size, 0.0 would produce length-zero edges at the center of mass.
"""
center_of_mass = np.mean(verts, axis=0)
shrunk_verts = [
np.average(
(v, center_of_mass), axis=0, weights=(scaling_factor, 1 - scaling_factor)
)
for v in verts
]
return shrunk_verts
def generate_offsets(num_meshes, scaling_factor=3):
"""
Generate vectors to displace vert segment membership symbols in a ring around the vert marker
"""
angular_dist = 2 * np.pi / num_meshes
return (
scaling_factor
* np.array((np.cos(angular_dist * x), 0, np.sin(angular_dist * x)))
for x in range(num_meshes)
)
# shape primitives --- use draw_shape()
def draw_sphere(position, scale=1):
return f".sphere {' '.join(str(x) for x in position)} {scale}\n"
def draw_cube(position, scale=1):
offset = np.array((0.605, 0.605, 0.605)) * scale
bottom_left = position - offset
upper_right = position + offset
return f".box {' '.join(str(x) for x in bottom_left)} {' '.join(str(x) for x in upper_right)}\n"
def draw_cone(position, scale=1):
offset = np.array((0, 0.5, 0)) * scale
base_center = position - offset
tip = position + offset
return f".cone {' '.join(str(x) for x in base_center)} {' '.join(str(x) for x in tip)} {scale}\n"
def draw_cyl(position, scale=1):
offset = np.array((0, 0.5, 0)) * scale
base_center = position - offset
tip = position + offset
return f".cylinder {' '.join(str(x) for x in base_center)} {' '.join(str(x) for x in tip)} {0.75 * scale}\n"
def draw_arrow(position, scale=1):
offset = np.array((0, 0.75, 0)) * scale
base = position - offset
tip = position + offset
return f".arrow {' '.join(str(x) for x in base)} {' '.join(str(x) for x in tip)} {0.6 * scale} {0.8 * scale} 0.5\n"
shapes = [draw_sphere, draw_cyl, draw_cube, draw_cone, draw_arrow]
def draw_shape(vert_position, map_offsets, group, scale=1):
"""
Draw the correct shape for a given tetra group
"""
offset_position = map_offsets[group] + vert_position
shape_idx = group % len(shapes)
return shapes[shape_idx](offset_position, scale)
def draw_fusion_marker(vert_coord, group_idx_pair, map_offsets, scale=1):
"""
Draw a thin black cylinder between two group symbols for a given vertex
"""
start_pos = vert_coord + map_offsets[group_idx_pair[0]]
end_pos = vert_coord + map_offsets[group_idx_pair[1]]
return f".cylinder {' '.join(str(x) for x in start_pos)} {' '.join(str(x) for x in end_pos)} {0.1 * scale}\n"
def get_colors(num_groups, color_scale=None):
plot_colors = "#2f357c,#b0799a,#e69b00,#355828,#6c5d9e,#bf3729,#e48171,#f5bb50,#9d9cd5,#17154f,#f6b3b0,#ada43b".split(",")
if "#" in color_scale:
plot_colors = color_scale.split(",")
assert all(
len(x) == 7 and x[0] == "#" for x in plot_colors
), "Enter comma-separated hex colors with #, no spaces"
else:
import matplotlib as mpl
try:
plot_colors = [mpl.colors.rgb2hex(x) for x in mpl.colormaps[color_scale].colors]
except KeyError:
print(f"{color_scale} is not a recognized matplotlib scale. Using the default scale.")
return plot_colors
def mesh_to_bild(
mesh,
filename,
colors,
map_offsets,
cyl_width=0.1,
jitter=0.2,
shrink=0.66,
rigid_range=1,
):
"""
Convert CryoSPARC mesh files into a series of BILD files for investigation.
"""
df = pl.DataFrame(
{
"cells": mesh["tm_cells"],
"segments": mesh["tm_segmask"],
"rigidity": mesh["tm_rweights"],
}
)
num_segments = len(set(mesh["tm_segmask"]))
max_rigidity = max(mesh["tm_rweights"])
psize = mesh["psize_A"]
box_offset = mesh["N"] / 2
mesh_points = [
psize * (np.array((box_offset, box_offset, box_offset)) + x)
for x in mesh["tm_points"]
]
already_drawn = []
combined_filename = filename.replace(".bild", "_combined.bild")
with open(combined_filename, "w") as all_f:
df_by_mesh = df.partition_by("segments", as_dict=True)
for group in range(num_segments):
group_filename = filename.replace(".bild", f"_g{group}.bild")
with open(group_filename, "w") as group_f:
file_list = [all_f, group_f]
# perform offsets at the BILD file level so that each tetra still has the correct
# (scaled) verts, in case that ever matters
if jitter:
group_offset = jitter * psize * random_three_vector()
else:
group_offset = (0, 0, 0)
group_offset = tuple(str(x) for x in group_offset)
write_multi(
file_list,
f".color {' '.join(str(x) for x in group_color(colors, group))}\n",
)
write_multi(file_list, f".translate {' '.join(group_offset)}\n")
sub_df = df_by_mesh[group]
# create edges
for face in sub_df.rows():
verts = face[0]
verts = tuple(mesh_points[x] for x in verts)
rigidity = face[2]
if shrink:
verts = shrink_tetra(verts, scaling_factor=shrink)
edges = combinations(verts, 2)
for edge in edges:
src_vert, dest_vert = edge
string_edge = paste_vectors(group, src_vert, dest_vert)
if string_edge in already_drawn:
continue
else:
already_drawn.append(string_edge)
write_multi(
file_list,
" ".join(
(
f".cylinder {' '.join(str(x) for x in src_vert)}", # start
f"{' '.join(str(x) for x in dest_vert)}", # end
f"{cyl_width * (1 + rigid_range * rigidity / max_rigidity)}\n", # radius
)
),
)
# remove the jitter so that the next group starts from offset (0, 0, 0)
write_multi(file_list, ".pop\n")
verts_filename = filename.replace(".bild", "_verts.bild")
with open(verts_filename, "w") as verts_f:
file_list = [all_f, verts_f]
# base verts transparent so you can see fusion segments through them
write_multi(file_list, ".transparency 0.5\n.color 0.7 0.7 0.7\n")
all_verts = tuple(tuple(x) for x in mesh_points)
unique_verts = set(all_verts)
for vert in unique_verts:
write_multi(file_list, f".sphere {' '.join(str(x) for x in vert)} 2\n")
# ChimeraX cannot display two transparent objects if they overlap/intersect,
# so even though it would be nice to have the tetra group symbols be transparent
# as well it's better to leave them opaque.
write_multi(file_list, ".transparency 0\n")
verts_by_group = {x: [] for x in range(num_segments)}
for tet_index, tet_verts in enumerate(mesh["tm_cells"]):
group_num = mesh["tm_segmask"][tet_index]
for vert_idx in tet_verts:
verts_by_group[group_num].append(all_verts[vert_idx])
verts_by_group = {k: tuple(set(v)) for k, v in verts_by_group.items()}
# write each tetra group's symbols in one go to avoid writing tons of unneccessary
# .color directives, which would bloat the BILD file.
for group_num in range(num_segments):
write_multi(
file_list,
f".color {' '.join(str(x) for x in group_color(colors, group_num))}\n",
)
for vert_position in verts_by_group[group_num]:
write_multi(
file_list, draw_shape(vert_position, map_offsets, group_num)
)
# to draw fusion connectors we need to switch to thinking about "For each vert, which groups"
vert_group_membership = {}
for group_num, vert_list in verts_by_group.items():
for vert_coord in vert_list:
if vert_coord not in vert_group_membership:
vert_group_membership[vert_coord] = set()
vert_group_membership[vert_coord].add(group_num)
assert sum(len(x) for x in vert_group_membership.values()) == sum(
len(x) for x in verts_by_group.values()
)
# now that we know we properly captured the groups for each vert, we can drop
# verts for which there is only one group, since they will definitely not need any
# fusion connectors
vert_group_membership = {
k: v for k, v in vert_group_membership.items() if len(v) > 1
}
write_multi(file_list, ".color 0 0 0\n")
for vert_coord, group_idxs in vert_group_membership.items():
group_idxs = list(group_idxs)
group_idxs.sort()
for group_idx_pair in combinations(group_idxs, 2):
if (
group_idx_pair in mesh["segfuses"]
# we no longer care whether A -> B or B -> A
or group_idx_pair[::-1] in mesh["segfuses"]
):
write_multi(
file_list,
draw_fusion_marker(
vert_coord,
group_idx_pair,
map_offsets=map_offsets,
scale=1,
),
)
def main(args):
if args.edge_width > 0.5:
print("Edge widths greater than 0.5 often produce poor results.")
with open(Path('~/instance-info.json').expanduser(), 'r') as f:
instance_info = json.load(f)
cs = CryoSPARC(**instance_info)
assert cs.test_connection()
project_number = args.project if "P" in args.project else f"P{args.project}"
job_number = args.job if "J" in args.job else f"J{args.job}"
project = cs.find_project(project_number)
job = project.find_job(job_number)
mesh_out = job.load_output("flex_mesh", slots=["mesh"])
with open(project.dir() / mesh_out["mesh/path"][0], "rb") as pkl:
mesh = pickle.load(pkl)
# we need all tetras to be assigned to a group, so None won't work
if mesh["tm_segmask"] is None:
mesh["tm_segmask"] = [0] * len(mesh["tm_cells"])
mesh_out_dir = Path().absolute() / f"tetra-mesh_{project_number}-{job_number}"
mesh_out_dir.mkdir(exist_ok=True)
mesh_out_filename = mesh_out_dir / f"{project_number}-{job_number}_mesh.bild"
num_groups = len(set(mesh["tm_segmask"]))
plot_colors = get_colors(num_groups=num_groups, color_scale=args.color_scale)
palette_filename = mesh_out_dir / f"{project_number}-{job_number}_palette.txt"
with open(palette_filename, "w") as f:
f.write("palette " + ":".join(plot_colors) + "\n")
print("for chimeraX: palette " + ":".join(plot_colors))
plot_colors = list(matplotlib.colors.to_rgb(x) for x in plot_colors)
map_offsets = list(generate_offsets(num_groups))
mesh_to_bild(
mesh,
filename=str(mesh_out_filename),
colors=plot_colors,
map_offsets=map_offsets,
cyl_width=args.edge_width,
jitter=args.jitter_mag,
shrink=args.tet_shrink,
rigid_range=args.rigid_range,
)
zip_filename = f"{project_number}-{job_number}_mesh-bild"
shutil.make_archive(
base_name=zip_filename,
format="zip",
root_dir=mesh_out_dir,
)
print(f"Done. Archive at {zip_filename}")
parser = argparse.ArgumentParser(
usage="Convert a CryoSPARC mesh into a .bild file for ChimeraX"
)
parser.add_argument("project", help="Project number")
parser.add_argument("job", help="Job number")
parser.add_argument(
"--jitter_mag",
help="Change the displacement of each tetra group. Default 0.2",
type=float,
default=0.2,
)
parser.add_argument(
"--edge-width",
help="Width of edges. Scaled by rigidity. Default 0.1",
type=float,
default=0.1,
)
parser.add_argument(
"--tet-shrink",
help="Shrink each tet toward its center of mass by this proportion. Default 0.66",
type=float,
default=0.66,
)
parser.add_argument(
"--rigid-range",
help="The most rigid segment will be (rigid-range * edge_width) times wider than the least rigid segment. 0 sets all edges to the same width. Default 1.0",
type=float,
default=1.0,
)
parser.add_argument(
"--color-scale",
help="Override default color scale. Give a matplotlib palette name (e.g., Dark2), or a comma-separated list of hex colors.",
type=str,
)
if __name__ == "__main__":
args = parser.parse_args()
main(args)