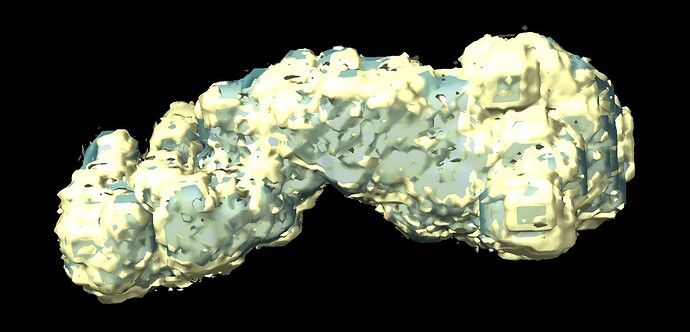
Hello!
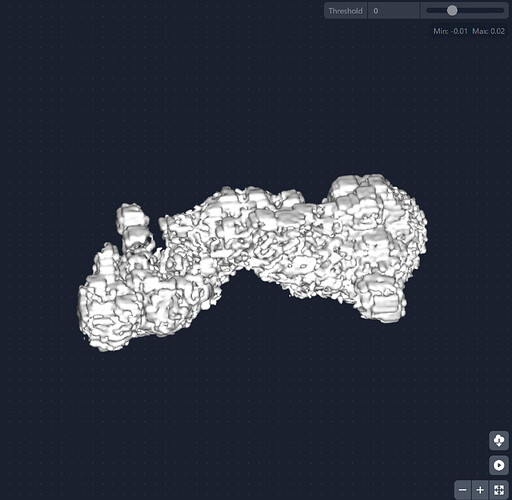
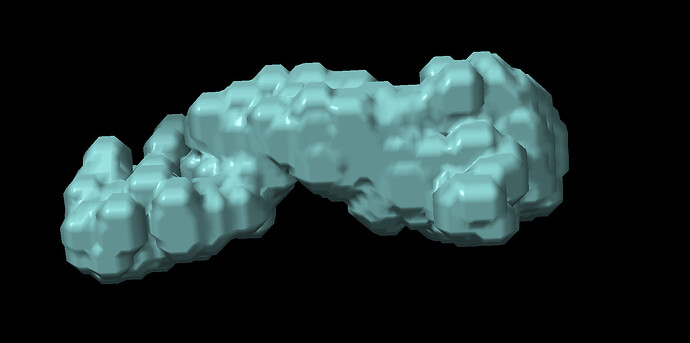
I am working on a small protein complex (~170 kDa) that has motions in a very small region (approximately 70 amino acids). The complex has C2 pseudo-symmetry and motion in this part can be seen after 3DVA. I tried running 3D-flex to get a better refinement with this region, but my resulting volume looks as if its been binarized. I’m pretty new to this, so I’m not sure if there is a step I’m missing in the data processing that comes after 3D reconstruction.
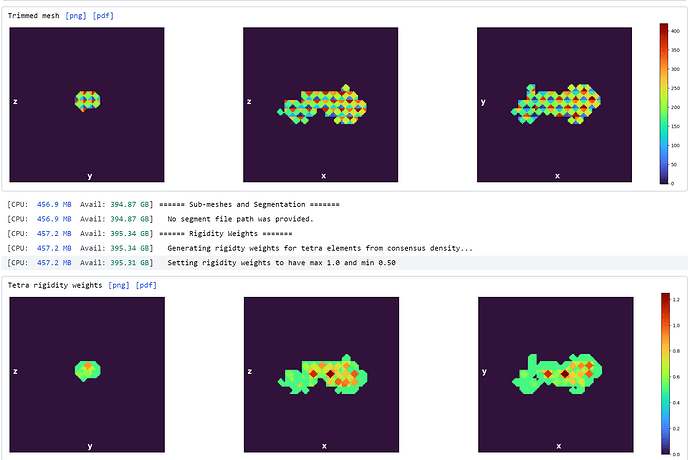
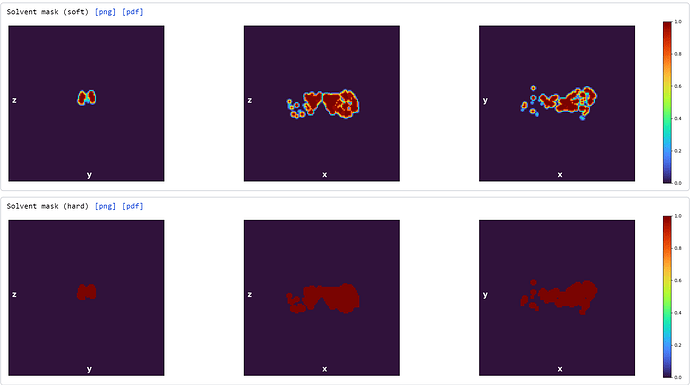
I’ve tried “loosening” the rigidity parameters in the mesh prep and training since my region of interest is so small as well as messing with the latent centering strength (still trying to find a good value for this). The reconstruction job that produced this volume has all default parameters.
Does anyone know why my resulting volume looks like this? Are there parameters I should be adjusting in the mesh prep/training steps to avoid this result?
Thank you so much in advance!