Hello everyone,
I’m having an error i cant understand
Traceback (most recent call last):
File “cryosparc_master/cryosparc_compute/run.py”, line 116, in cryosparc_master.cryosparc_compute.run.main
File “cryosparc_master/cryosparc_compute/jobs/flex_refine/run_meshprep.py”, line 167, in cryosparc_master.cryosparc_compute.jobs.flex_refine.run_meshprep.run
File “cryosparc_master/cryosparc_compute/jobs/flex_refine/tetraseg.py”, line 105, in cryosparc_master.cryosparc_compute.jobs.flex_refine.tetraseg.segment_and_fuse_tetra
AssertionError: All voxels in mask should have been assigned to a region but were not.
to do my segmentation i’ve used the 3d flex prep job output map at a threshold of 0.3,
and also i created a mask from the same volume at the same threshold.
So i’m quite confused as to what i did wrong
I have 3 segmentation and i try two different connection tree
A > B wich show the previous error and i tried
C > A > B and it changed the error to
Traceback (most recent call last):
File “cryosparc_master/cryosparc_compute/run.py”, line 116, in cryosparc_master.cryosparc_compute.run.main
File “cryosparc_master/cryosparc_compute/jobs/flex_refine/run_meshprep.py”, line 149, in cryosparc_master.cryosparc_compute.jobs.flex_refine.run_meshprep.run
ValueError: too many values to unpack (expected 2)
-
did you segment the volume from 3D Flex Data Prep or somewhere else ?
-
Does the syntax A>B, C>B make sense based on the segments in Chimera ?
seems the issue is with this input in the .seq file or connecting the mesh.
Hello @Mark-A-Nakasone,
I did the segmentation from 3D flex data Prep and i used the same treshold I used for the 3D mesh prep job.
A>B, C>B does make sense but i would like not to link C and B
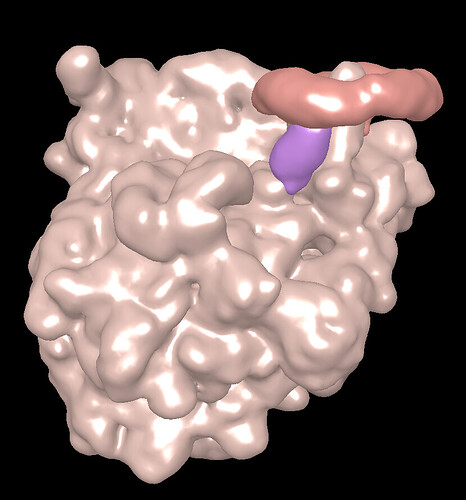
A is purple, B is redish and C is brownish
B and C are indeed in contact however, it’s two separate molecule and we know from our data that red is higly flexible.
A is from the same molecule as B, however it is less flexible and fairly strongly attached to the brown part, so it being linked to it makes sense at it’s bases.
That volume seems to have a lot detail for 3D Flex.
It would just be a matter of what is the ID of A, B C. In UCSF ChimeraX it would be somewhat random e.g. A=435, B=435, and C=321. The segger tool works in both Chimera and ChimeraX, but in ChimeraX it could save the output (.seg file). Differently. Which Chimera are you using ?
https://www.youtube.com/watch?v=KmWyjVSkiic was good example
I’m using Chimera and i checked the number for A, B and C multiple time they are correct and i changed them everytime i retried the segmentation to make sure they match the new one.
I’ve had a look at the tutorial already could not help me.
A=1581, B=2094, C=2095
the volume is from the 3D data prep job with a pixel size of 2.12 A and i used a Nu refinement volume as the input volume of the job.
suggests to play with
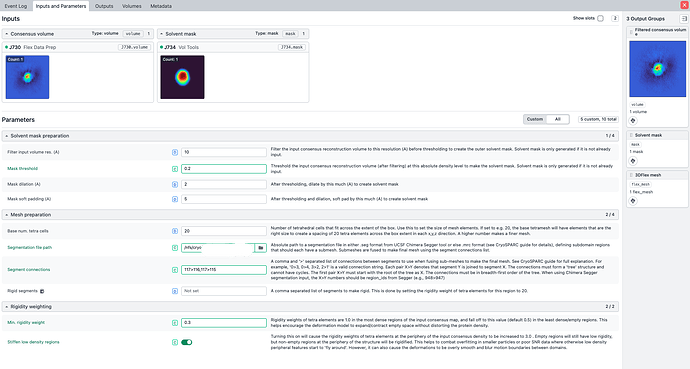
“Mask dilation (A)” = maybe go to ~8
“Mask soft padding (A)” = maybe go to 12-32
That could work around that error.
At 2.12A I usually filter the volume a bit for 3D Flex, anywhere from 6-24A depending on how much movement.
What is the exact syntax for your “Segment connections” ?
Hi @Mark-A-Nakasone,
Sorry for the late answer,
My intial syntax was “1581 > 2094”, i just tested " 2095 > 1581, 1581 > 2094" and it works.
However,
I’m unhappy with that solution because when i look at the mesh, i see that inside a single vertices there are density of A and C. but A and C are linked together at the base of A, not throughout the mass.
is there a solution to go around that ?
should i make a segmentation for just the base of A and and do “C > base, base > A, A > B”
@Yannlefrancois glad that it is at least making the mesh. From my understanding this should not be a major issue. You can always place with number of tetracells, but likely don’t have to. For 3D Flex training attaching the per-particle data from 3DVA can be helpful.
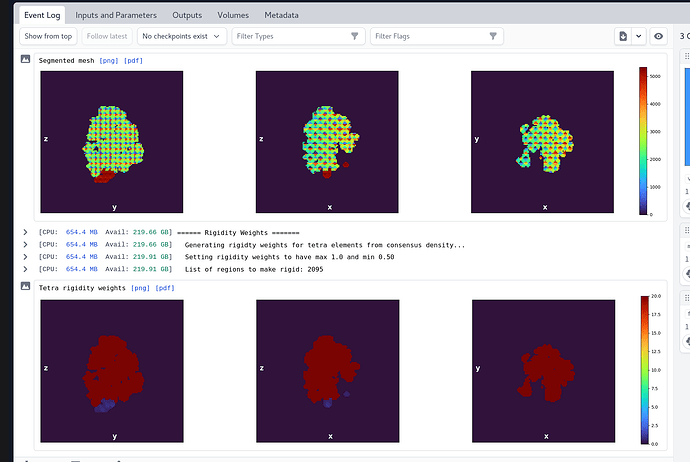
I maybe cannot see is here, but are the tetracells segmented, e.g. on the events page you see different regions ?
@Mark-A-Nakasone
I think it is separated in different region as they have different color on the event page.
I was reluctant to play with tetracells size because of computation time.
I’ve done a 3D variability analysis but with a mask on the flexible region not on the whole particule.
Can it still work if i want to do 3d flex on the whole particule or do i need to redo my 3DVA without the mask ?
The particle meta data from 3DVA should be able to go with any segmented or non-segmented mesh. You just want to ensure the particles have the same linage going to 3DVA and 3D Flex Data prep - everything should match.
I am not sure how local/focus refine is computed, but in general your segmented volumes appear very small in respect to to total volume. Could be poor/low signal with such a small region, but give it a try.
It’s not just poor/low signal volume, it’s just the relative size of the different part of my complex.
I’ll try to add the 3DVA information to my 3D flex.
Thanks a lot for your help ^^
1 Like