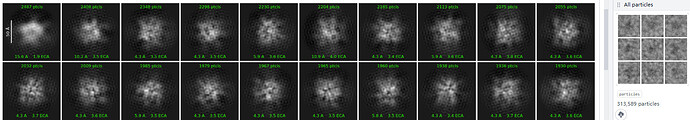
I‘m currently working on a small protein about 120kd, and its structural part is probably only 50kd
this is the 2D result, pretty weird
dose anyone what causes this? maybe anyway to make it better?
thanks a lot!
How big is your box? It looks too small for this particle
but in the right part, I feel like the box is enough?
the solid part of my particle is estimated to be around 80A
this is superresolution mic, and is crop to 1/2 in motion corretion, now the real pixel size is 0.512A
my box size is 256pixel (131A, that’s 1.64 times of the estimated diameter)
Definitely too small
You have to consider CTF delocalization, which displaces information by an absolute amount that doesn’t depend on the size of your particle (just on defocus and electron wavelength).
So for a ribosome 1.6x would be fine, but for something this size you need a larger box relative to particle size. Consult Rosenthal & Henderson JMB 2003 for a more detailed discussion
never thought about that, thanks a lot!
For a more methodological examination, check out this manuscript as well:
https://www.biorxiv.org/content/10.1101/2020.08.23.263707v1
Some of it was published in another paper, so sadly I don’t think it will ever be formally published. ![]()
does that mean i should nit make my sample too dense? if i make it too dense, the box would contain two paricles. is ithat ok?
It’s generally a good idea to have some “empty ice” around particles, but it’s not always possible; and for some samples, you can force orientations which otherwise are disfavoured by pushing concentration that little bit higher. ![]()