Dear community,
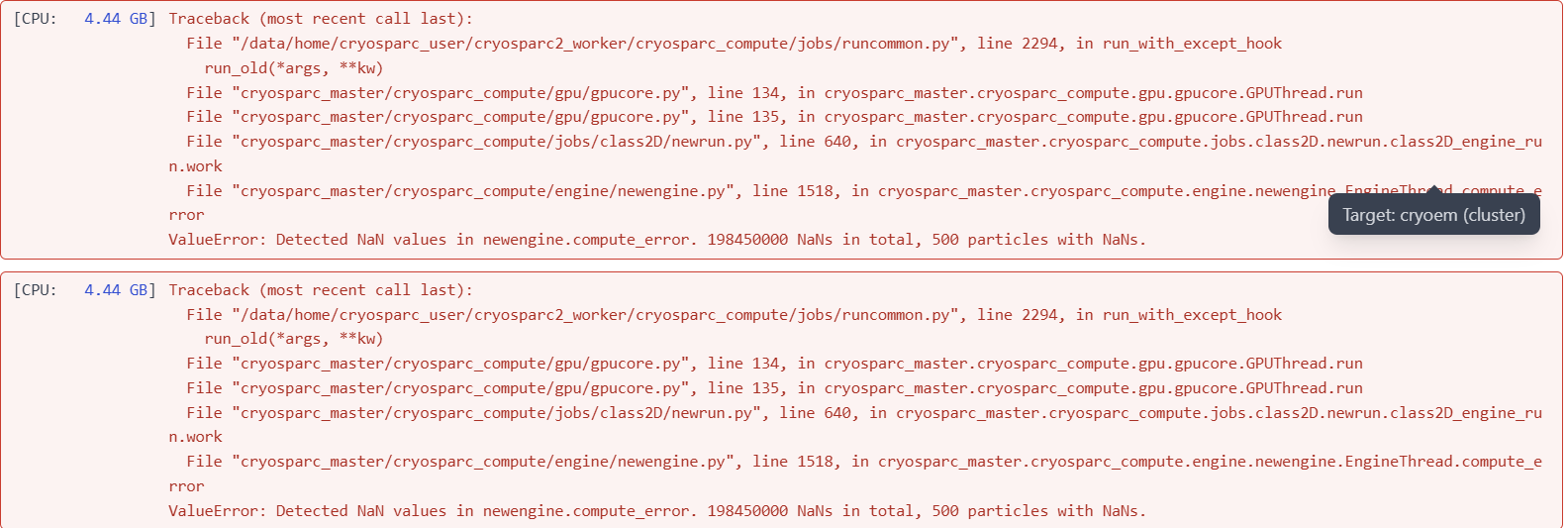
My 2D classification was terminated by error messages related to detected NaN values, even if the “check particle” job confirmed no corrupted particles with NaN values. Can anyone please inform me how to proceed?
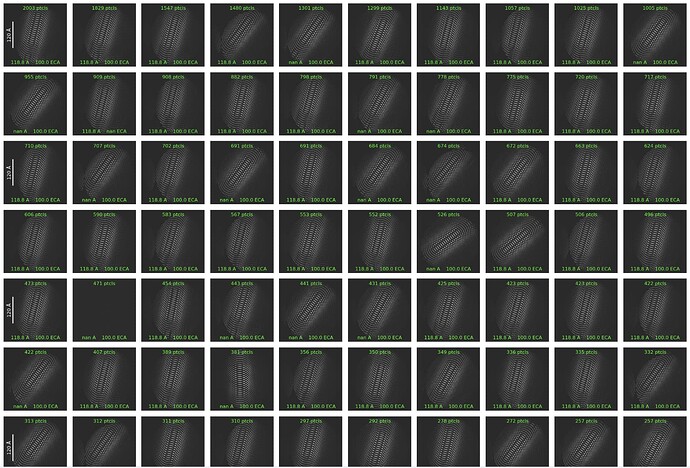
Please see the screenshot of the error message and the weird result of iteration 0.
Hi @dcui91,
These classes look very odd. Have you checked whether there was an error during import, e.g. wrong Cs or wrong pixel size?
Do you expect your particles to look like this, based on what you see in the micrographs?
Cheers
Oli
Hello Oli,
Thanks for the reply. I checked the import, and I didn’t see anything wrong. The desired particles do not look like this. The 2D classification of small subsets of the particles (around 30K) looks normal 2D average.
I just checked through all the 2D classification jobs related to this dataset. Only 2 jobs are successful (30K and 120K particles respectively), and all other jobs failed no matter the total number of particles. Some show NaN-related error messages at the end of iteration 1, and some show the error message at around 10th iteration. This is my first time seeing error messages like this since I used cryosparc, and I really have no idea what leads to that issue.
Also, heterogenuous refinement works fine.
—Da
Hi Da - you can try running “Check for Corrupt Particles” on the input stack - perhaps there is a small group of bad/corrupted particles that are throwing it off?
Hi Oli,
The weird thing is that all particles come from the “check for coruppted particles” job, which says no corruptions detected.
—Da
@dcui91 For the Check for corrupt particles job, please can you post the output of the command
csprojectid="P99"; csjobid="J199" # replace with actual project and job ids of relevant job
cryosparcm cli "get_job('$csprojectid', '$csjobid', 'type', 'version', 'params_spec', 'status')"
Hello,
Where can I see the command of the job?
By the way, the problem was solved somehow, without any change of the input from last time. Everything just went back to normal somehow…
The commands would have to be run on a Linux shell on the CryoSPARC master computer.