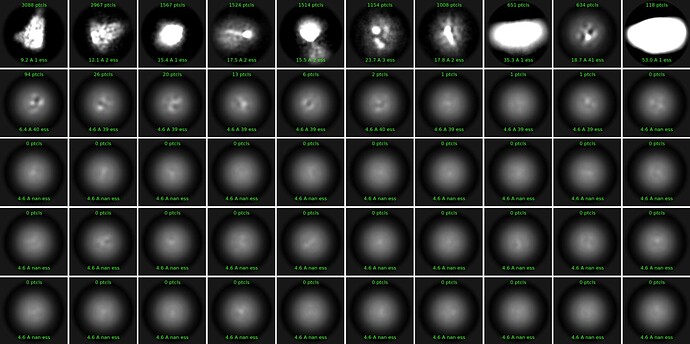
Hello,
We collected single particle cryoEM data on a coronavirus spike ecto-domain on a Krios/Falcon4/Selectris configuration at a national facility. Cryosparc-live spit out a very nice 3A resolution map with all the expected features during data collection. However, when we imported the movies into our home cluster and performed 2D classification after the usual motion correction and CTF estimation, we ran into an unusual problem. The class averages look extremely bright and show few secondary features. A snapshot (J1420) is attached. Then, we switched on the Force Max over poses/shifts tab and also tried Align filament classes vertically (BETA) ON. This improved the 2D classification a little bit but it is nowhere near the quality we have seen for other datasets. Any advice on how to fix this apparent brightness and sort of smearing issue is appreciated. Thank you.
Saif