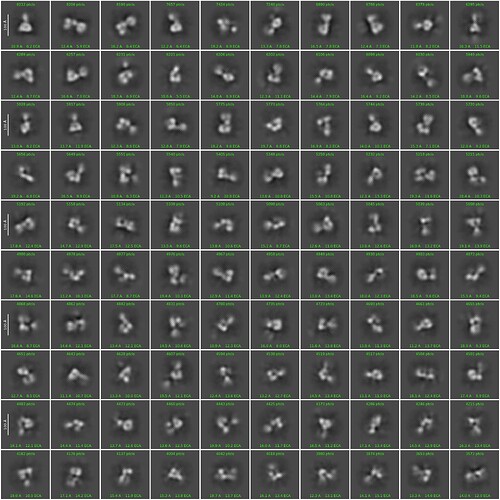
Hello, everyone. The size of my protein is 150KDa. More than 5000 pieces of data were collected on 300KV electron microscope. The diameter of extracted particles was 150A and the box size was 256px. After three rounds of 2D, I still feel a little unclear. Is there any way to improve it?Thank you so much for your help.
mask diameter:180A
Batchsize per class:200
iteration to anneal sigma:30
I would also like to add that the box size is currently bin2.
Can you have a try using the settings of “Force max over poses/shifts=OFF” and increase “batchsize per class” to 500?
I have tryed to increase “batchsize per class” to 500, so I would to try “Force max over poses/shifts=OFF” next.Thanks!
Hi niu,
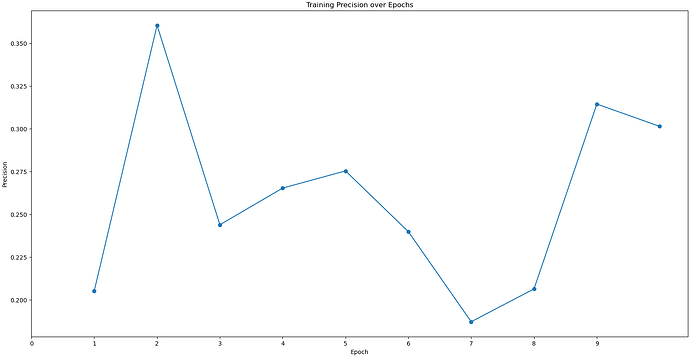
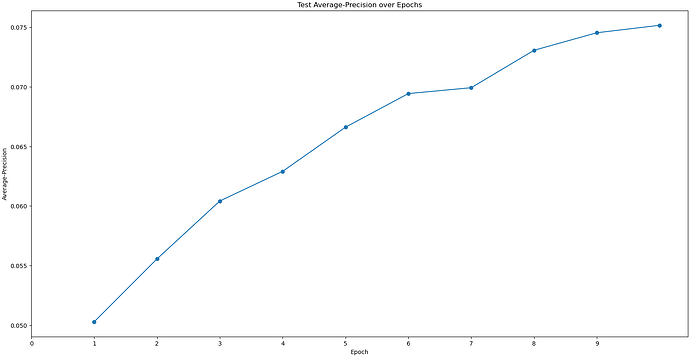
I would like to ask that I used"Topaz Train"to train a model, and using about 500,000 particles, but the result of this train seems to be not very good, could you please help to check it?Thank you very much.
What’s the pixel size ? Depending on the value, and regarding the size of your particle of interest, I’d suggest to tweak the box size to be ~ 1.5x your particle size in pixels. How are your micrographs looking? If noisy or carbon picks, I can also suggest to try micrographs denoiser (Beta).
Following up on @KevinM 's point, a larger box size may help. 1.5 to 2x the longest diameter of your protein is usually recommended for smaller proteins. I’d also increase your online EM iterations to 30 and increase your batch size to 300. I’d also keep using the mask, but maybe try it at 190-200 Å since it looks like you’re still getting some of the protein clipped off. Force max on is helpful, but you do need to increase your batch size and online EM iterations to help improve.
Better particle picking may also help, since it looks like you still have quite a bit of noise (or maybe thick ice?), so playing with Topaz or Deep Picker may be helpful. With Topaz, you need to make sure you have very good particles that are being used to train on, which is likely why your Topaz training job stats don’t look great.
Hi,now I want to use Topaz Denoise, and “Micrograph” I used output files of “Curate Exposures” as Input files. I wanted to use output files from" Import Movie" as “training Micrograph” input files, but the “eer” format were not available. I have only used output files of “Curate Exposures” as “Micrograph” input files and “training Micrograph” is empty. Is this “Denoise Model” credible? I’m still new to this, so I haven’t tried it yet.
Hi @Linqian,
If you are using a version of CryoSPARC that is v4.5 or newer and used Patch Motion Correction to motion correct your micrographs, we would gently recommend to try our Micrograph Denoiser. This can be found in the Exposure Curation portion of the Job Builder tab. More information on how the job works and what you should do if you run into any issues can be found here.
Best,
Kye
I’d second trying the CryoSPARC denoiser.
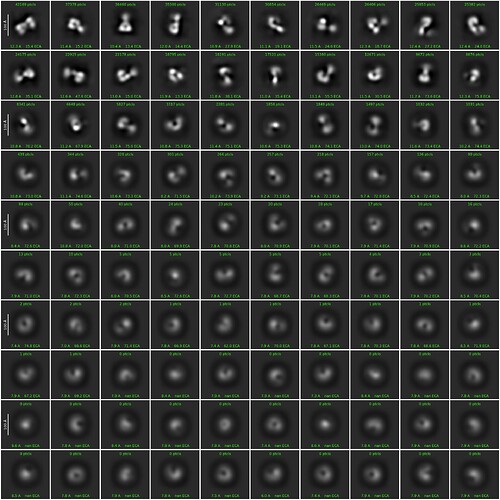
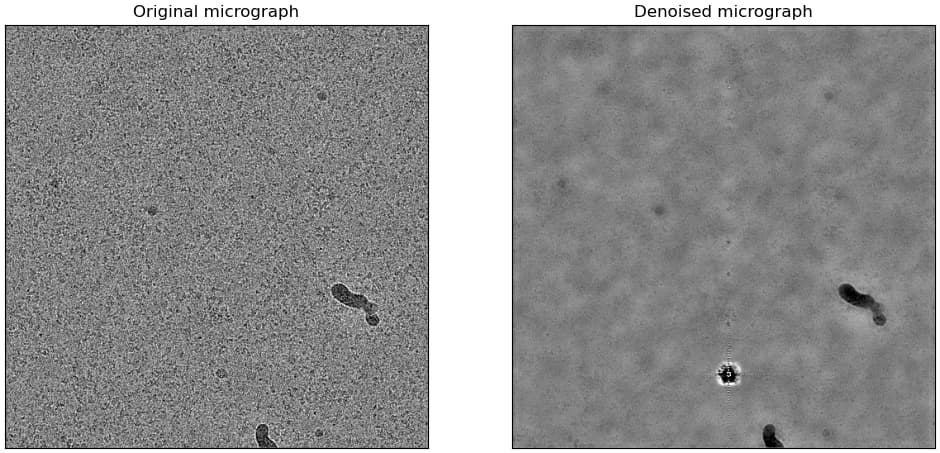
However, with one caveat - don’t use the pretrained model. Train a model from your data. I’ve had really poor results with the pretrained model for a wide variety of data. An example using apoferritin, although this is a bit of an edge case because of some other tests I was running:
Pretrained model:
Self-trained model:
…
Other than that, for something so small, I’d try a short acquisition at significantly higher magnification as the particles on those micrographs look very… bitty. Broken, (partially) aggregated, generally a bit too heterogeneous. If the particles are the same size (in Angstrom) then you might have a slightly easier time picking as they’ll be effectively larger and a lot easier to identify the quality of the prep. Then you can go back and adjust.
Thanks for your suggestion, I will try it.
Thank you for your advice. Thank you