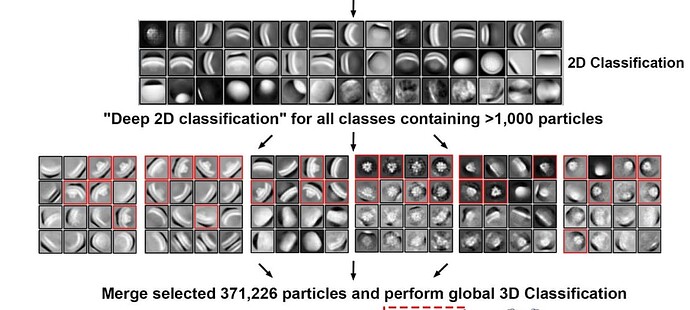
I would be interested in seeing this workflow supported as well, possibly in conjunction with 2D classification without alignments (2D classification without alignment?).
For now, is there a way to implement this workflow with cryosparc tools? E.g.
for Job X:
if class >1000 particles:
select class:
run 2D classification on selection
Is that possible with CS tools…? Perhaps optionally implementing reference-based 2D class selection for the 2D class selection in the subclassifications?
Cheers
Oli
EDIT:
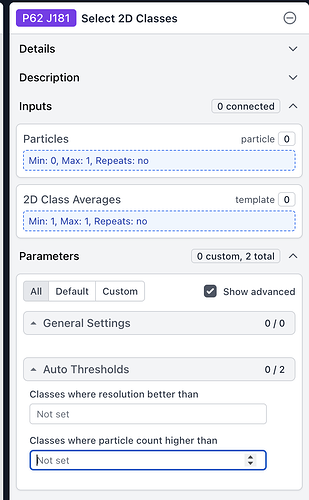
An easy way to facilitate a “manual” version of this approach would be to have a “split outputs by class” option in Select 2D, in addition to auto thresholds: