You are very much correct, high symmetry targets would be a pain! I have noted these applications. I also took note of your other request:
EDIT: For purposes like this, it would be helpful if Apply Trajectories generated a few plots of the particle trajectories on the micrographs, as RBMC itself does, so that one could double-check that it is behaving as expected.
Unsure about the “job process terminated abnormally” message, please send logs if it happens again. Its possible there was a resource issue due to a specific mic with a high number of particles or two mics being processed on the same GPU. It might be necessary to limit 1 mic per GPU by setting the oversubscription value to greater than the GPUs vRAM.
1 Like
Hi @kstachowski,
I have a question about this workflow.
In the righthand (incorrect) branch, using sym expand plus VAT, how would you expect Apply Traj. to handle the missing trajectories?
I ask because (in my ignorance) I had already tried this incorrect approach, and it did not give an error and it output the expected number of particle stacks (the sym expanded number, twice the number of the parent “whole-vault” RBMC job from where the trajectories were applied).
So in that case does it just output without applying any motion correction, or takes trajectories from a parent patch motion job? It should probably give an error or warning if a trajectory is missing for a given UID, right?
Cheers
Oli
Hi @olibclarke,
After some discussions with other folks on the team, it seems I was wrong about a couple of items above. The workflow you did (right hand column), should work correctly with no issues. The confusion on my end is what information Apply Trajs is using to do the RBMC. To eliminate any future confusion from others, I will be editing my post above to cleanup the incorrect info.
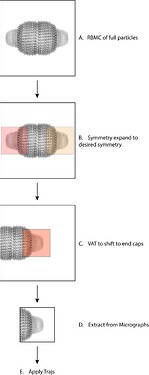
So, here is a correct description of the workflow and whats happening under the hood.
Step A generates a host of information in the particle.cs file including a set of info in the motion slot. This includes motion/path and motion/idx. For each micrograph, a .npy file is generated containing all of the motion trajectories for each particle. In the case of this workflow, this is the trajectory of the entire vault complex. In each .npy file are the trajectories of all particles on each micrograph and each particle is assigned a motion trajectory id (motion/idx).
Step B will duplicate all particle information for each symmetry mate except for generating a new particle UID and documenting from which particle the symmetry mate was generated (sym_expand/src_uid). The duplicated information includes both motion/path and motion/idx which are important for use later on. This step also changes the expanded particles’ alignments3D/pose as part of the symmetry expansion, such that both caps are coincident.
Step C will change all of the particles alignments3D/shift so that the cap is located at the center of the box.
Step D is used specifically to recenter the particles locations using the location/center(x/y)_frac and the new shifts produced from Step C. Apply Trajs does not recenter the particles; this step is likely required for correct results. You should also be fine with extracting at a smaller box size here although I did not test this on the downstream step.
Step E is where all of this info comes together. In this example, lets consider particle 1 is symmetry expanded to 1a and 1b. Apply Trajs will take particle 1a and 1b and apply the assigned trajectory (motion/idx in motion/path) coming from particle 1 (full vault complex) to each of the sub particles at each of those sub particle locations (location/center_(x/y)_frac). Therefore, the particle UIDs are not actually used (and this is where i went wrong). The output subparticles should now be RBMC’d using the full particles info.
2 Likes
Thank you Kye - that detailed explanation is very helpful!!
One thought - in cases like this, it would be helpful in the initial RBMC job to be able to calculate the trajectories, without generating/writing the motion corrected particles (because we are going to do that in the next step for subparticles with Apply Traj.). This would save some diskspace and presumably some computation?
Cheers
Oli
Hi @olibclarke,
Currently, trajectories are only saved once per-particle motion correction is performed. During hyperparameter optimization and empirical doseweighting, there are trajectories that are calculated but these are not saved, especially since only a small subset of the data is seen during these two steps.
BUT, you make a good point and I have noted this, as it would really help out in these types of cases. It will definitely save disk space, but note though, that the trajectory optimization is often more expensive than making the corrected particle images (it can vary with hardware, box size, and other data-dependent factors), so the performance differential may be marginal.
Best,
Kye
2 Likes
Is it safe to set the Fourier crop to say 2 or 4 pixels, to minimize data written at this stage? I assume that is the last step so won’t affect the trajectories.
The only other thing that might be an issue is that RBMC by default recenters particles prior to motion correction (presumably prior to calculating the trajectories).
Presumably VAT takes shifts into account, so I guess this doesn’t matter for the workflow as described - as presumably they are either there and taken into account, or reset to zero during RBMC with recentering?
Hi @olibclarke,
Is it safe to set the Fourier crop to say 2 or 4 pixels, to minimize data written at this stage? I assume that is the last step so won’t affect the trajectories.
In Step A or D?
The only other thing that might be an issue is that RBMC by default recenters particles prior to motion correction (presumably prior to calculating the trajectories).
Presumably VAT takes shifts into account, so I guess this doesn’t matter for the workflow as described - as presumably they are either there and taken into account, or reset to zero during RBMC with recentering?
Is this is regards to a future feature where we have an “only write trajectories” option or this workflow?
In Step A or D?
In step A - the full particle RBMC job - to avoid writing out large full particles which we may not need if the goal is subparticle reconstruction.
Is this is regards to a future feature where we have an “only write trajectories” option or this workflow?
No the current workflow - what I was wondering is whether (because RBMC recenters particles by default) it is neccessary to perform a refinement of the RBMC full particles, and use that for VAT, or whether one can simply use the RBMC input coordinates & volume for VAT (in order to get the coords for subparticle correction in Apply Traj.).
Since F-cropping is one of the last steps, this theoretically should be fine because the trajectories are calculated from particles using the movie pixel size and cropped at the end. CPU/GPU limitations should still be considered since the full box size and movie pixel size are used.
Your second point might warrant against the above because depending on the magnitude of the shifts present for the full particle, recentering might change the particles’ location by a large amount. If you have done a few previous extractions and refinements, you might be okay.
I might recommend first doing RBMC on a subset of all the particles, cropping to 128 or 256 px and doing a homogeneous reconstruct and judging based on that if it would be viable to skip a homogenous reconstruct and refinement with the full particle stack.
1 Like
Hi Kye,
Trying this now, and have encountered one issue - when RBMC is run with manually input hyperparameters, it does not generate an output hyperparameters slot to use with Apply trajectories.
In this case we run the dose weighting with different hyperparameters from the trajectory estimation. If I just provide the output micrographs from the final RBMC run in the “movies” slot, will it get the trajectories & dose weighting params from there? Or is it necessary to explicitly provide the hyperparameter slot?
EDIT: I guess the trajectories are encoded in the particles.motion slot, and the supplied hyperparameters only control dose weighting?
Step D is used specifically to recenter the particles locations using the location/center(x/y)_frac and the new shifts produced from Step C. Apply Trajs does not recenter the particles; this step is likely required for correct results. You should also be fine with extracting at a smaller box size here although I did not test this on the downstream step.
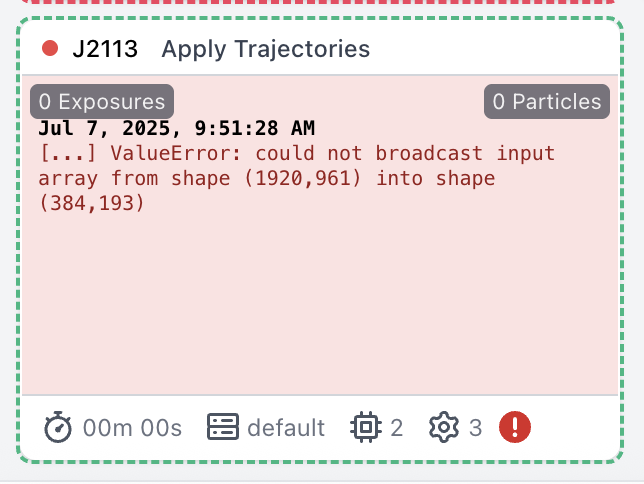
If I try extracting at a smaller box size, I get this error:
The original box size during the full-particle RBMC was 960px, F-cropped to 700; the box size during the extraction after VAT was 384px (no F-crop). So I guess I need to perform the extraction at the original box size (although perhaps it will allow an arbitrary F-crop to save space…?)
Using the whole box size at the post-VAT extraction step has the downside that a lot of particles will be discarded if they are anywhere close to the edge of the micrograph. It would be useful, in cases like this, to have an option to recenter using aligned shifts in Apply Traj, which would obviate the need for the post-VAT extraction.
EDIT:
Using the full box size before Apply Traj still gives the same error. I think it is complaining about the output box size in Apply Traj. Indeed, if I set the output box size in Apply Traj. to the “movie-scale” box size of the whole vault (1920px), it does run, but that will fill up my disk rather quickly…
Hey @olibclarke,
I guess the trajectories are encoded in the particles.motion slot, and the supplied hyperparameters only control dose weighting?
Correct!
As for your most recent post, that error is for the EDWs. They only work for the same box size, movie pixel size, and frame count that they were computed for, so you will need one additional step before apply trajs where you extract the subparticles at the boxsize you want to use and then do RBMC to just compute the EDWs. I should have caught this earlier.
1 Like
Hmm - so in that step, for the subparticle EDW estimation, you would use as inputs the extracted particles (now centered on the caps), a reconstruction of said particles, and a matching mask?
Will this then require tweaking of the hyperparameters during EDW to avoid the previously-documented late-frame-upweighting bug in RBMC?
Hey @olibclarke,
Hmm - so in that step, for the subparticle EDW estimation, you would use as inputs the extracted particles (now centered on the caps), a reconstruction of said particles, and a matching mask?
Yeah, I believe this would be the best course of action. You might be able to get away with a homogenous reconstruction instead of doing a full refinement.
Will this then require tweaking of the hyperparameters during EDW to avoid the previously-documented late-frame-upweighting bug in RBMC?
I would recommend connecting the hyperparams output from the full particles if you spent time tweaking to correct for the late-frame upweighting; especially since the trajectories were derived from those hyperparameters. Make sure to remove the EDWs at the LLI before launching the job or just type the values into the override slots.
So in summary, you should have:
- RBMC to get trajs of the whole particle
- Symm Expand + VAT + Re-extract to get the subparticles
- Reconstruction of caps
- RBMC using full particle hyperparams and particles/volume/mask from step 3 to get EDWs of subparticles
- Apply trajs with whole particle trajs and subparticle particles and EDWs
2 Likes
Hi Kye,
I’m afraid it still doesn’t work - it still complains there is a mismatch, even after I redo the EDW estimation from this particle set.
Could it be because the original patch motion job was performed with Fcrop=1/2, whereas the RBMC trajectories were at the original, unaltered pixel size?
EDIT:
… nevermind, I’m an idiot - because there is no Fcrop option in Apply Traj., I need to extract at the box size in “movie pixels” (768px) in order to match what was going on in the EDW job, and then downsample in a separate job. Then it works.
EDIT2:
Is the first plot in Apply Traj. the EDWs? If so the scaling seems a little weird, and it is hard to interpret:
Yeah, that plot is currently not useful. Also, thanks for working through this. It is providing lots of insights to us to help make these workflow possible. We appreciate your efforts! Please let me know if you see any improvements.
1 Like
Hi Kye,
It worked insofar as it improved compared to without RBMC, however not yet better than direct “full” RBMC of the subparticles after symmetry expansion and local refinement - so far they seem pretty comparable. However of course it is going to vary case-by-case, and this has been a very helpful exchange to get my head around the workflow - thank you for your patience!
Cheers
Oli
1 Like