Hi @liz,
Thanks for your patience, I had to spend some time exploring how to do this effectively within CS. I was able to make a workflow to find the portal on the Calicivirus VP2 dataset, EMPIAR-10193.
After Homogeneous Refinement of your virus you can do the following.
-
Symmetry expand the final particle stack with
Isymmetry. -
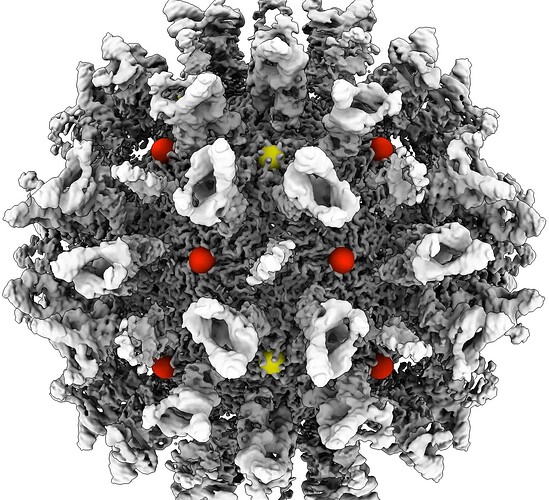
Use ChimeraX to locate the vertex of which the particles and volume will be shifted to the center of the box. I am unsure if this will apply to your dataset, but for EMPIAR-10193 there were two possible vertices where the particle could be shifted too (red and yellow), of which the the red vertex was the correct one.
To find the coordinates of this position, place a marker on the surface, reposition the marker if needed using the “Move” mode in the “Markers” panel, and use
measure center #{marker}to obtain coordinates in Å.
-
Back in cryosparc, connect the volume and symmetry expanded particles to a Volume Alignment Tools (VAT) job.
-
Use the 3D-coordinates output by the measure command in chimeraX as input for the VAT parameter “3D-coordinates of new center” and use ‘A’ for Angstroms. In my case, this was “366.67,419.19,559.45 A”.
-
Download your shifted volume, and find the right location for a mask using the same protocol from step 3.
-
Using the coordinates from step #6, I followed @olibclarke’s tutorial on creating cylindrical masks. I generated a mask around the area where the portal should be.
- I did not gaussian filter the volume, I lowpass filtered it in CS.
-
Import the mask into your workspace using the Import Volumes job.
-
Use a Volume Tools job to lowpass filter the mask to 15 Å and pad with 30 pixels. Although these are the values that worked for me, I didn’t test many – others will probably work well!
-
Next, re-extract the shifted particles using Extract From Micrographs.
- For the dataset that I tested this workflow on, I re-extracted at a box size of 384 px and Fourier cropped to 300 px.
- I then did a Homogeneous Reconstruction to ensure the putative viral portal was in the center of the box and that the mask I created fit over that region as well.
- Connect the particle output of the homogeneous reconstruction to a 3D-classification job and the cylindrical mask as the focus mask. For parameter choices, I set the following:
- Number of classes: 80
- Filter resolution: 6
- Initial structure lowpass: 15
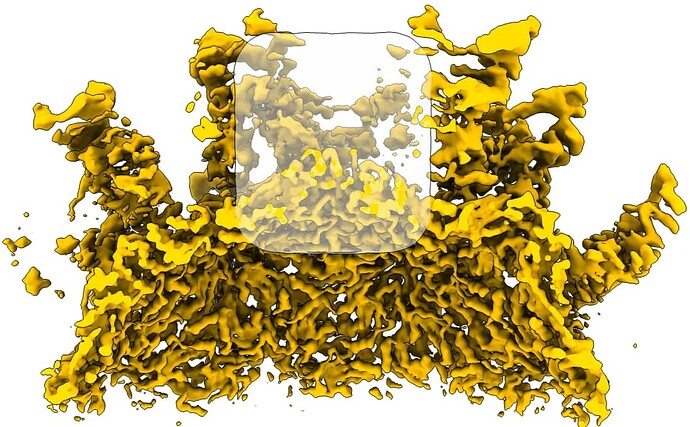
This job then had one class which has the portal density clearly visible:
I hope this helps and please reach out if you have any further questions.
Best,
Kye