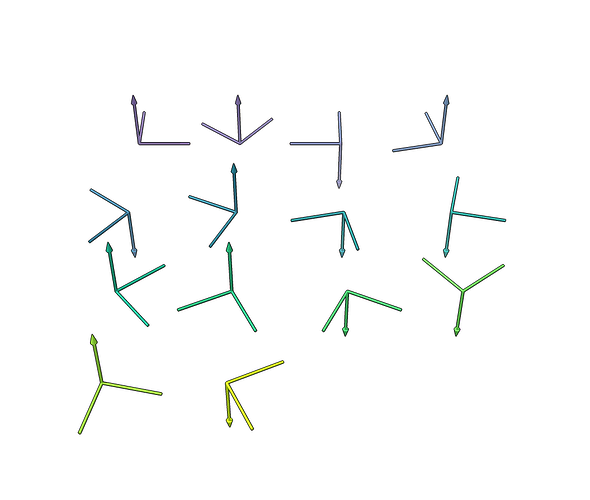Hi @Adwaith99!
At the bottom of this reply is a script you can use (you’ll need cryosparc tools, matplotlib, and scipy) to generate BILD files like this one to inspect the symmetry operators. If you hover over the axes ChimeraX will display which index corresponds to that rotation.
This image is the result of setting symmetry_order to D7.
I’m still not sure I personally quite understand the usefulness of this workflow, and want to make sure you saw the caveat I mentioned above – not all particles in the same symmetry expanded index are in identical pose with regards to symmetry-breaking features.
Now for your question about how the index order is generated. The short answer is that the indices are the same every time, but unfortunately there’s not currently a straightforward way to know which is which without looking manually.
Script
from cryosparc.tools import CryoSPARC
import json
import numpy as np
from pathlib import Path
from scipy.spatial.transform import Rotation as R
import matplotlib.pyplot as plt
with open(Path('~/instance-info.json').expanduser(), 'r') as f:
instance_info = json.load(f)
cs = CryoSPARC(**instance_info)
assert cs.test_connection()
project_number = "P337"
workspace_number = "W7"
symmetry_order = "D7"
lane_name = "cryoem9"
# any job with a particles output will do - we're just using this
# dataset to avoid having to create all the fields manually
arbitrary_job_with_particles = "J68"
project = cs.find_project(project_number)
particles = project.find_job(arbitrary_job_with_particles).load_output("particles")
particles = particles.take([0])
particles["alignments3D/pose"][0] = [0., 0., 0.]
particles["alignments3D/shift"][0] = [0., 0.]
fake_particles_uid = project.save_external_result(
workspace_uid = workspace_number,
dataset = particles,
type = "particle",
)
symexp_job = project.create_job(
workspace_uid = workspace_number,
type = "sym_expand",
connections = {
"particles": (fake_particles_uid, "particle")
},
params = {
"sym_symmetry": symmetry_order
}
)
symexp_job.queue(lane_name)
symexp_job.wait_for_done()
exp_particle = symexp_job.load_output("particles")
colors = plt.get_cmap("viridis")(np.linspace(0.1, 0.9, len(exp_particle)))
with open(f"{symmetry_order}.bild", "w") as f:
arrows = np.array([
[1, 0, 0],
[0, 1, 0],
[0, 0, 1]
])
axes_per_bild_row = int(np.round(np.sqrt(len(exp_particle.rows()))))
for row in exp_particle.rows():
idx = row["sym_expand/idx"]
x_translation = 2 * (idx % axes_per_bild_row)
y_translation = -2 * (idx // axes_per_bild_row)
f.write(".pop\n")
f.write(f".translate {x_translation} {y_translation} 0\n")
f.write(f".color {' '.join(str(x) for x in colors[idx][:-1])}\n")
rotation = R.from_rotvec(row["alignments3D/pose"])
pointing_directions = rotation.apply(arrows)
f.write(f".note idx{str(idx)}\n")
for axis in pointing_directions:
f.write(f".cylinder 0 0 0 {' '.join(str(x) for x in axis)} 0.02\n")
arrow_base = pointing_directions[2]
arrow_point = arrow_base * 1.2
f.write(f".cone {' '.join(str(x) for x in arrow_base)} {' '.join(str(x) for x in arrow_point)} 0.05\n")