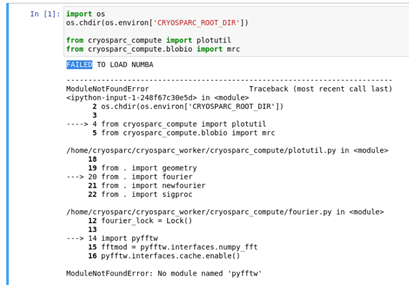
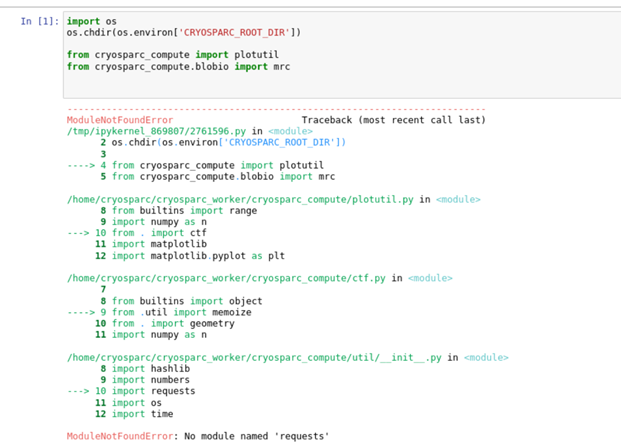
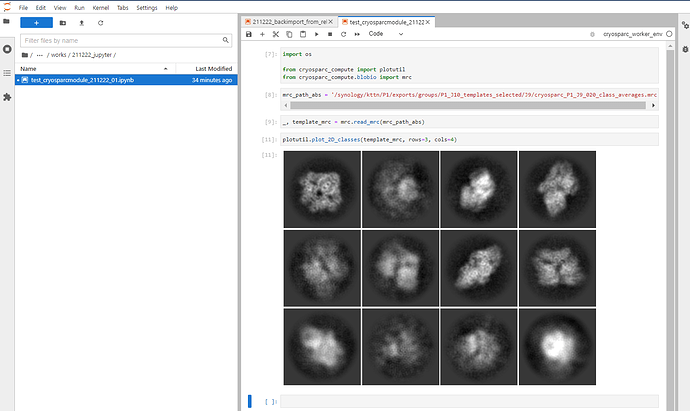
I’m using the cryosparc modules in jupyter-lab in the following way.
Please replace the directory paths for your environment.
(1) Activate worker conda environment
[cryosparcuser@gx03 /amorphous/cryosparcuser/cryosparc/cryosparc_worker]
> source deps/anaconda/envs/cryosparc_worker_env/bin/activate
(2) Install ipykernel module
(cryosparc_worker_env)
[cryosparcuser@gx03 /amorphous/cryosparcuser/cryosparc/cryosparc_worker]
> conda install ipykernel
(3) Install the kernelspec for the cryosparc worker python environment
(cryosparc_worker_env)
[cryosparcuser@gx03 /amorphous/cryosparcuser/cryosparc/cryosparc_worker]
> ipython kernel install --user --name cryosparc_worker_env --display-name cryosparc_worker_env
Installed kernelspec cryosparc_worker_env in /usr/people/cryosparcuser/.local/share/jupyter/kernels/cryosparc_worker_env
Then logout/re-login to reset environment variables.
(4) Install and activate a fresh anaconda environment
(If you laready have your own anaconda, then skip this step and just activate your virtual environment.)
Get an installer of the latest anaconda and execute it.
[cryosparcuser@gx03 ~]
> ./Anaconda3-2021.11-Linux-x86_64.sh
Tempolarily activate the anaconda environment.
[cryosparcuser@gx03 ~]
> eval "$(/usr/people/cryosparcuser/anaconda3/bin/conda shell.bash hook)"
(5) Make sure the installed cryosparc kernelspec is recognized
(base)
[cryosparcuser@gx03 ~]
> jupyter kernelspec list
Available kernels:
cryosparc_worker_env /usr/people/cryosparcuser/.local/share/jupyter/kernels/cryosparc_worker_env
python3 /usr/people/cryosparcuser/anaconda3/share/jupyter/kernels/python3
(6) Launch jupyter-lab
(base)
[13:35:01 cryosparcuser@gx03 ~]
> PYTHONPATH=/amorphous/cryosparcuser/cryosparc/cryosparc_worker jupyter-lab --ip=0.0.0.0 --no-browser --notebook-dir='/' --port=8888
Specifying the PYTHONPATH is important.
Access the jupyter-lab server with your web blowser.
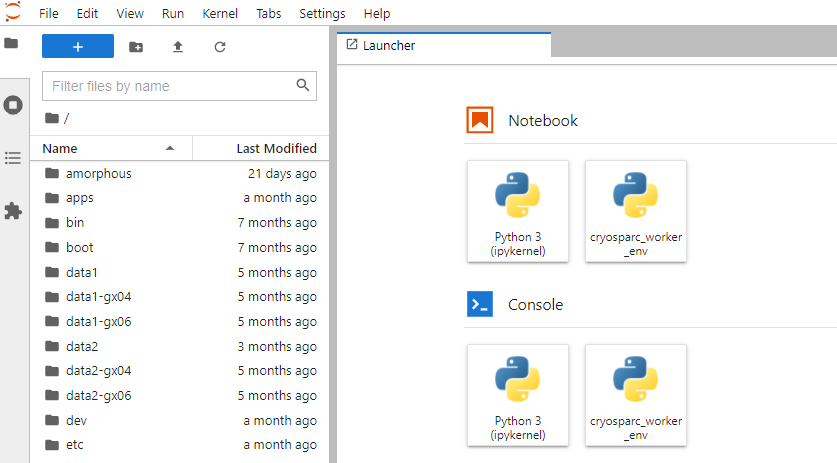
Then you can see the installed kernelspec as an available kernel for Notebook/Console.
Launch a new notebook with the cryosparc_worker_env kernel selected, then you can use the cryosparc module.
Hope this helps.