Hi @Ahn! In addition to the guidance above, I want to note that if you’re importing just particle pick locations, your star file has neither raw data nor pose data. So you’re right that you should turn on both of Ignore raw data and Ignore pose data.

You’ll also need to fill out the Micrograph Parameter Overrides section with the appropriate information, since it’s not present in your STAR file.
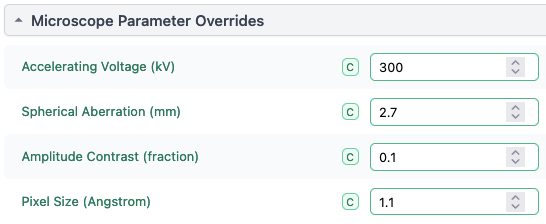
(those settings should be replaced with whatever is appropriate for your images, of course)
The most important of these is the Pixel Size (Angstrom). Your pick locations from crYOLO are in pixels X/Y, but CryoSPARC uses a convention of what fraction along the micrograph in X/Y. The pixel size is needed to convert between these two, and that’s likely the field that the import job is complaining about being missing.
And finally, as far as the micrograph names, what you’re trying to get to match are
- The names of the micrographs you used for crYOLO
- The names of the input micrographs for this job
Since you used the motion-corrected micrographs for crYOLO try plugging the output of Patch Motion Correction or Patch CTF Estimation into this job’s Source Exposures input and leaving all “Length of … to cut” as 0 and see if that works!