Dear @Ahn,
I would check again your .star file. Does your .star file have data_optics header present in the header such as below? If not, there are some microscope param overrides to set when you are building Import Particle Stack.
data_optics
loop_
_rlnOpticsGroup #1
_rlnOpticsGroupName #2
_rlnAmplitudeContrast #3
_rlnBeamTiltX #4
_rlnBeamTiltY #5
_rlnSphericalAberration #6
_rlnVoltage #7
_rlnImagePixelSize #8
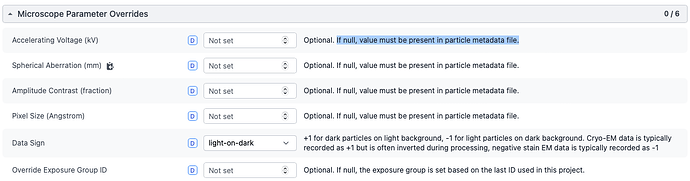
You are getting “AssertionError: Could not import star file: too many fields missing.”, because microscope parameters (voltage, Cs, amplitude contrast, and pixel size) need to be present to import particle stacks (please see the screenshot below). If these parameters are not in your .star, I’d provide them in the import job.
I also noticed that you did not provide the Source Exposures for input. To preserve the particle locations, you would need the source micrographs as the input when you’re importing the particle stack.
Additionally, editing the .star with sed to truncate the micrograph names is not needed. When you are building Import Particle Stack, you can tweak the lengths of both the input exposure and the query rlnMicrographName.
For your case, I would cut 4 characters (.mrc) from input exposure file name suffix and 31 characters (_patch_aligned_doseweighted.mrc) from rlnMicrographName base name suffix to have everything end with _Fractions.
I’d suggest having a look at the guide if you have not already done so:
Best,
Kookjoo