Hi @AndreGraca,
You can do this by first downloading the movie_blob .cs file output from the exposures_rejected output result group of the Manually Curate Exposures job.
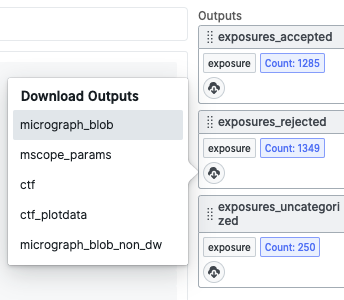
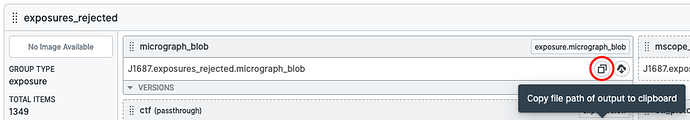
You can also just get the path to the .cs file on the master node by navigating to the Outputs tab and pressing the “copy path” button on the movie_blob output:
Once you have the .cs file, you can then open it up using the instructions from our guide on manipulating .cs files here:
https://guide.cryosparc.com/processing-data/manipulating-.cs-files-created-by-cryosparc
You can then find the filenames, and either delete the files manually, or use python to do this for you.
For example, open a shell on the master node and run cryosparcm icli to start an interactive python session, then run the following:
from cryosparc2_compute import dataset
exposure_dset = dataset.Dataset() #initialize the dataset object
dataset_path = "<path_to_cs_file_here>"
exposure_dset.from_file(dataset_path) #load the .cs file
exposure_dset.data['movie_blob/path'] #will print out a sample of all the values in this field
# the following will write all the filenames to a text file that
# can be piped to a unix delete command
with open("exposures_to_delete.txt", 'w') as openfile:
for file_to_delete in exposure_dset.data['movie_blob/path']:
openfile.write(file_to_delete + '\n')
#the following will delete the files sequentially
import os
for file_to_delete in exposure_dset.data['movie_blob/path']:
try:
os.remove(file_to_delete)
except:
print("Unable to delete {}".format(file_to_delete))
continue