@GYADAV correct, but whichever .cs file is first has priority. So you will want the _particles file first to get the new parameters from that job.
Thank you both Oli and Daniel.
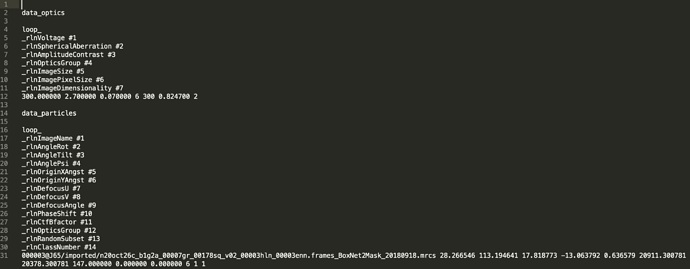
I used following command and it generated star file. Do I need P6_J50_passthrough_particles.cs? What information it have?
csparc2star.py /data/shared/Gaya/Wt_Fv_nano_Purdue/P6/J50/cryosparc_P6_J50_024_particles.cs cryosparc_P6_J50_024_particles.star
I used following command to convert selected particles stacks, but somehow it does not carry the defocus values in the star file. Does I need to provide any specific option to carry the defocus values?
csparc2star.py particles_selected.cs particles_selected.star
Thank you
have you tried using the passthrough file as an additional input cs file?
Thank you,
It works good with passthrough file.
Hello,
Somehow I cannot use my imported mask in the refinement. I tried to drag it, but not working.
What might be wrong?
thanks
Gaya
Did you make sure to set “Type of volume being imported: mask” and not map?
Thank you
Yes, I make that mistake.
Gaya
I converted a cryosparc_P22_J87_008_particles.cs to cryosparc_P22_J87_008_particles.star . When I do a Bayesian Polish job in Relion, I get the following error:
ERROR:
StackHelper::splitByMicrographName: rlnMicrographName missing from MetaDataTable .
I initially picked the particles using WARP. I have exported micrograph list from WARP and input it to the Micrographs (from MotionCorr) tab on the Bayesian polishing job. What is the best way to tackle this error?
@acabezas Please post your entire command. The mostly likely reason the micrograph names are not present is that a passthrough file wasn’t provided.
thank you for the response. the Relion command is the following;
which relion_motion_refine --i P22_J87_passthrough_and_particles.star --f PostProcess/job008/postprocess.star --corr_mic part2_pinkprotein_star.star --first_frame 1 --last_frame -1 --o Polish/job022/ --min_p 10000 --eval_frac 0.5 --align_frac 0.5 --params3 --only_do_unfinished --j 16 --pipeline_control Polish/job022/
The csparc2star.py command. You probably need to include the passthrough file, and then edit the star file to point to your Relion MotionCorr output.
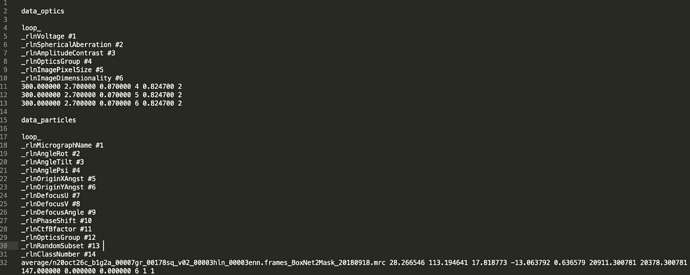
I included the passthrough_particles.cs in the csparc2star.py command. The csparc2star.py command I ran was the following;
python csparc2star.py cryosparc_P22_J87_008_particles.cs P22_J87_passthrough_particles.cs P22_J87_iter008_withpassthrough.star
It then resulted in the following STAR file;
I initially picked particles using WARP and processed the data up until a Non-uniform Refinement in Cryosparc. The error Relion gives me is -> StackHelper::splitByMicrographName: rlnMicrographName missing from MetaDataTable .
Is there a way I can run csparc2star.py command to include the rlnMircrographName in the output star file? If not, do I have to manually add it and edit the column such as in the following screenshot where I replaced _rlnImageName #1 with _rlnMicrographName #1?
Thank you for your assistance on this matter.
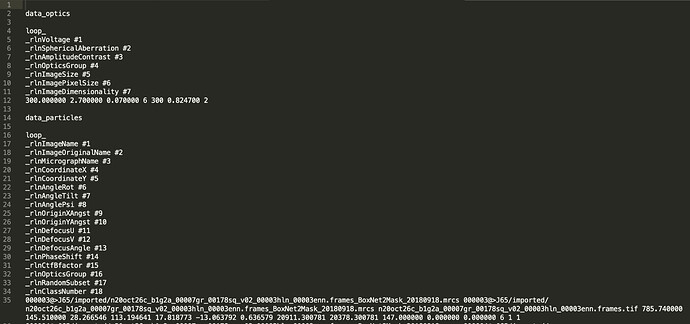
You need the micrograph names and the coordinates to extract particles. If you export cryosparc particles that have linked locations, or if you use --copy-micrograph-coordinates with the imported .star file that contains coordinates, then those fields will be preserved.
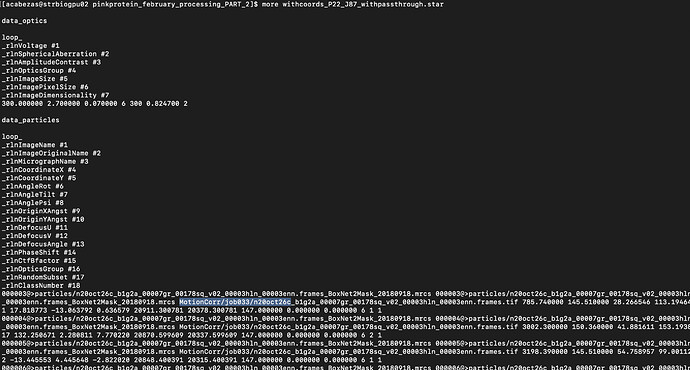
Thank you Daniel, --copy-micrograph-coordinates worked and the particles.star file contains the missing fields. Screenshot of that;
However, attempting to run a Bayesian Polishing job with this new particles.star file contains a new error.
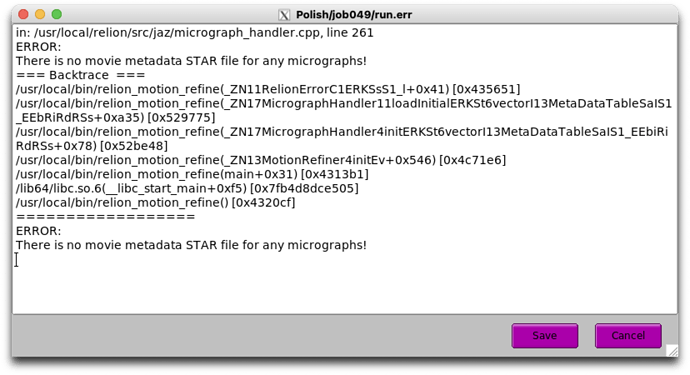
ERROR:
There is no movie metadata STAR file for any micrographs!
My micrographs.star file, which I got from from the “Export Micrograph List” option in WARP looks like this;
I’d appreciate any input on how to tackle this error.
-Andres
You need the movies themselves to run particle polishing, as well as the motion correction data captured by Relion when you run a MotionCorr job (doesn’t matter if MotionCor2 or Relion’s version is used, but you have to run it from Relion). Then the rlnMicrographName for the particles needs to be the micrographs for which Relion has motion data.
If you already have the Relion MotionCorr output, you probably just need to do one more search-and-replace in your particles .star file. You also need a PostProcess output as well but it sounds like you already have that part.
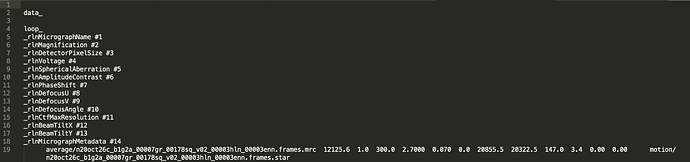
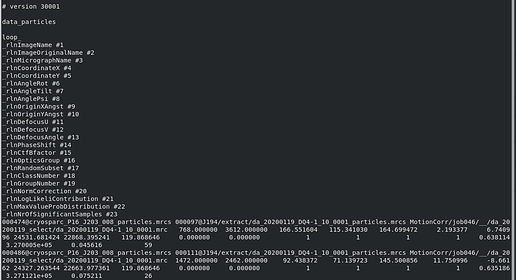
Here’s an example data.star from Refine3D that I then used for Polish. The rlnImageName is a converted cryoSPARC jobs, the rlnMicrographName has been edited to point to a MotionCorr output.
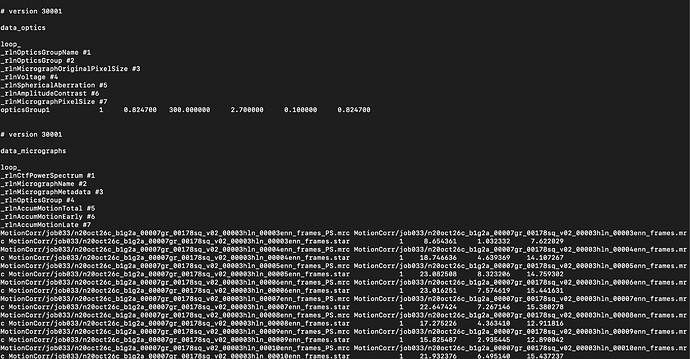
I ran Motion Correction in Relion using the imported Movies. Below is a screenshot of that MotionCorr output .star file.
I did the search-and-replace in my particles.star file (converted from .cs to .star using pyem ; after following the GitHub instructions for particles picked outside of cryosparc - in this case using WARP particle picker) to have rlnMicrographName #3 point to MotionCorr/job033/ like you suggested in your most recent reply.
Yet, I still get the same error : There is to movie metadata STAR file for any micrographs!
Any ideas on how to fix this or what data is missing? Thank you for your continued support.
Looks like you changed the file names to the movies, not the output micrographs like in my example screenshot. Also, always double check the path is valid from your Relion project directory, by changing into that directory and using ls <path>.
@DanielAsarnow
Is it possible for your csparc2star.py to extract the beam tilt and the beam trefoil parameters in Relion’s format from e.g., NU Refinement of cryosparc jobs? Maybe somebody already asked this kind of question? Thanks!